
Kadipiro virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Seadornavirus
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
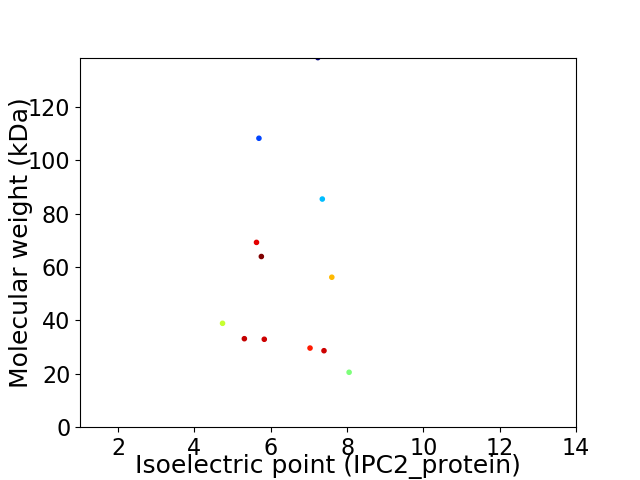
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9YWP6|Q9YWP6_9REOV Vp8 OS=Kadipiro virus OX=104580 PE=4 SV=1
MM1 pKa = 7.57SNIHH5 pKa = 6.96CYY7 pKa = 10.71GSDD10 pKa = 3.19PSEE13 pKa = 3.75GHH15 pKa = 5.19FCIYY19 pKa = 10.3RR20 pKa = 11.84FDD22 pKa = 3.73EE23 pKa = 4.74GQPFTVKK30 pKa = 10.24FRR32 pKa = 11.84QKK34 pKa = 11.03GKK36 pKa = 8.66IQHH39 pKa = 6.51IEE41 pKa = 3.92LEE43 pKa = 4.09NSIGHH48 pKa = 6.4NGAINYY54 pKa = 9.5GSIFDD59 pKa = 4.17TIKK62 pKa = 10.46YY63 pKa = 5.94FTSIVGLTLINNNMTIDD80 pKa = 4.02LNSVAVDD87 pKa = 3.06NRR89 pKa = 11.84FRR91 pKa = 11.84GNATMVALKK100 pKa = 10.85CNVLGNLFLKK110 pKa = 10.67KK111 pKa = 9.87IVSSDD116 pKa = 3.29LQLTLKK122 pKa = 10.54FYY124 pKa = 10.46YY125 pKa = 10.14NKK127 pKa = 10.16QIRR130 pKa = 11.84YY131 pKa = 9.22DD132 pKa = 3.72VFAGMYY138 pKa = 10.57GLINLRR144 pKa = 11.84NSKK147 pKa = 10.61FNDD150 pKa = 2.77NAYY153 pKa = 10.51NYY155 pKa = 10.55GIIIEE160 pKa = 4.1QCYY163 pKa = 9.2PIKK166 pKa = 10.29DD167 pKa = 3.56LPMNKK172 pKa = 8.87ILAGLKK178 pKa = 10.33ALMLFHH184 pKa = 7.03EE185 pKa = 5.01HH186 pKa = 6.16TNCIHH191 pKa = 6.91GDD193 pKa = 3.96CNPSNIMCDD202 pKa = 2.74KK203 pKa = 9.19MGNVKK208 pKa = 9.99IVDD211 pKa = 3.76PASLVTRR218 pKa = 11.84VVTYY222 pKa = 10.47INEE225 pKa = 4.24YY226 pKa = 10.8YY227 pKa = 10.58KK228 pKa = 11.0DD229 pKa = 3.92LTPKK233 pKa = 10.39SEE235 pKa = 3.85VGAYY239 pKa = 10.09LLSCFEE245 pKa = 4.27IVSDD249 pKa = 4.56LRR251 pKa = 11.84KK252 pKa = 10.11LPLEE256 pKa = 4.35KK257 pKa = 10.18IFIKK261 pKa = 10.29RR262 pKa = 11.84GYY264 pKa = 10.22LGLPEE269 pKa = 4.22YY270 pKa = 11.08ADD272 pKa = 4.27DD273 pKa = 4.77GGINAIDD280 pKa = 4.3FLTSLQDD287 pKa = 3.45GLTTHH292 pKa = 6.96SDD294 pKa = 3.29LMSAIAGVPFDD305 pKa = 4.99GLLANNEE312 pKa = 4.37YY313 pKa = 10.21IDD315 pKa = 6.08DD316 pKa = 4.48DD317 pKa = 5.67LSDD320 pKa = 3.89NEE322 pKa = 4.82CNVNLDD328 pKa = 3.56AGIDD332 pKa = 3.48NLEE335 pKa = 4.11FNNLDD340 pKa = 3.39VDD342 pKa = 4.73DD343 pKa = 5.38SDD345 pKa = 5.54SEE347 pKa = 4.3
MM1 pKa = 7.57SNIHH5 pKa = 6.96CYY7 pKa = 10.71GSDD10 pKa = 3.19PSEE13 pKa = 3.75GHH15 pKa = 5.19FCIYY19 pKa = 10.3RR20 pKa = 11.84FDD22 pKa = 3.73EE23 pKa = 4.74GQPFTVKK30 pKa = 10.24FRR32 pKa = 11.84QKK34 pKa = 11.03GKK36 pKa = 8.66IQHH39 pKa = 6.51IEE41 pKa = 3.92LEE43 pKa = 4.09NSIGHH48 pKa = 6.4NGAINYY54 pKa = 9.5GSIFDD59 pKa = 4.17TIKK62 pKa = 10.46YY63 pKa = 5.94FTSIVGLTLINNNMTIDD80 pKa = 4.02LNSVAVDD87 pKa = 3.06NRR89 pKa = 11.84FRR91 pKa = 11.84GNATMVALKK100 pKa = 10.85CNVLGNLFLKK110 pKa = 10.67KK111 pKa = 9.87IVSSDD116 pKa = 3.29LQLTLKK122 pKa = 10.54FYY124 pKa = 10.46YY125 pKa = 10.14NKK127 pKa = 10.16QIRR130 pKa = 11.84YY131 pKa = 9.22DD132 pKa = 3.72VFAGMYY138 pKa = 10.57GLINLRR144 pKa = 11.84NSKK147 pKa = 10.61FNDD150 pKa = 2.77NAYY153 pKa = 10.51NYY155 pKa = 10.55GIIIEE160 pKa = 4.1QCYY163 pKa = 9.2PIKK166 pKa = 10.29DD167 pKa = 3.56LPMNKK172 pKa = 8.87ILAGLKK178 pKa = 10.33ALMLFHH184 pKa = 7.03EE185 pKa = 5.01HH186 pKa = 6.16TNCIHH191 pKa = 6.91GDD193 pKa = 3.96CNPSNIMCDD202 pKa = 2.74KK203 pKa = 9.19MGNVKK208 pKa = 9.99IVDD211 pKa = 3.76PASLVTRR218 pKa = 11.84VVTYY222 pKa = 10.47INEE225 pKa = 4.24YY226 pKa = 10.8YY227 pKa = 10.58KK228 pKa = 11.0DD229 pKa = 3.92LTPKK233 pKa = 10.39SEE235 pKa = 3.85VGAYY239 pKa = 10.09LLSCFEE245 pKa = 4.27IVSDD249 pKa = 4.56LRR251 pKa = 11.84KK252 pKa = 10.11LPLEE256 pKa = 4.35KK257 pKa = 10.18IFIKK261 pKa = 10.29RR262 pKa = 11.84GYY264 pKa = 10.22LGLPEE269 pKa = 4.22YY270 pKa = 11.08ADD272 pKa = 4.27DD273 pKa = 4.77GGINAIDD280 pKa = 4.3FLTSLQDD287 pKa = 3.45GLTTHH292 pKa = 6.96SDD294 pKa = 3.29LMSAIAGVPFDD305 pKa = 4.99GLLANNEE312 pKa = 4.37YY313 pKa = 10.21IDD315 pKa = 6.08DD316 pKa = 4.48DD317 pKa = 5.67LSDD320 pKa = 3.89NEE322 pKa = 4.82CNVNLDD328 pKa = 3.56AGIDD332 pKa = 3.48NLEE335 pKa = 4.11FNNLDD340 pKa = 3.39VDD342 pKa = 4.73DD343 pKa = 5.38SDD345 pKa = 5.54SEE347 pKa = 4.3
Molecular weight: 38.94 kDa
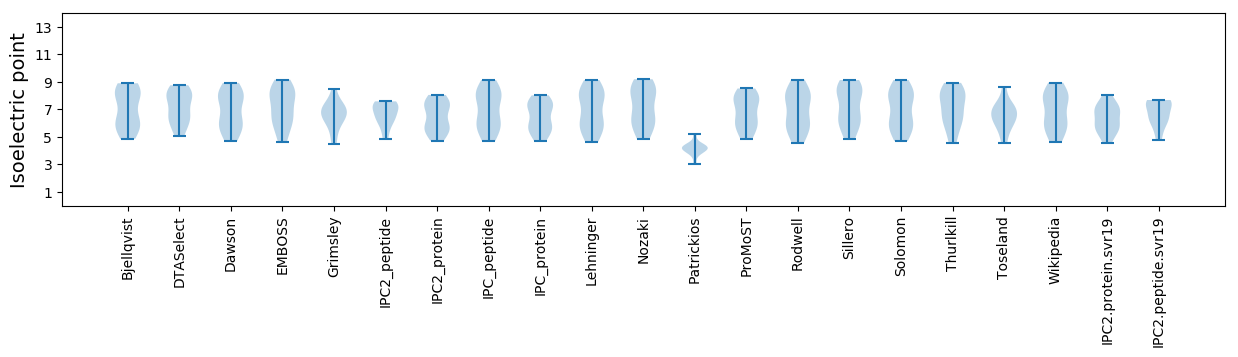
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9YYS1|Q9YYS1_9REOV Vp12 OS=Kadipiro virus OX=104580 PE=4 SV=1
MM1 pKa = 7.61SNARR5 pKa = 11.84YY6 pKa = 10.52NMISAVSASGCKK18 pKa = 9.02NDD20 pKa = 5.41KK21 pKa = 9.72PFCVHH26 pKa = 6.18KK27 pKa = 10.23VVAVIVIIVSSGITVVNQIGQRR49 pKa = 11.84KK50 pKa = 9.27LSTDD54 pKa = 3.39DD55 pKa = 3.91YY56 pKa = 11.18FVLSLVCDD64 pKa = 5.21AISNIIVVGLSVVGMTASSKK84 pKa = 9.13TKK86 pKa = 10.33QVLTDD91 pKa = 3.42GGEE94 pKa = 3.95NAIEE98 pKa = 4.13LNHH101 pKa = 5.4MPKK104 pKa = 10.3RR105 pKa = 11.84IVKK108 pKa = 10.16KK109 pKa = 10.41LLKK112 pKa = 10.32EE113 pKa = 3.81EE114 pKa = 4.39AKK116 pKa = 10.99NGVDD120 pKa = 4.03FVKK123 pKa = 10.91SIGKK127 pKa = 8.91DD128 pKa = 3.12LKK130 pKa = 11.61SNDD133 pKa = 3.66NKK135 pKa = 10.2PDD137 pKa = 3.49PSAPPKK143 pKa = 10.14YY144 pKa = 10.19EE145 pKa = 4.68KK146 pKa = 9.66IDD148 pKa = 3.41EE149 pKa = 4.27SFEE152 pKa = 4.34LKK154 pKa = 10.62SEE156 pKa = 4.19FAGKK160 pKa = 10.16SLITDD165 pKa = 3.07NRR167 pKa = 11.84TGQSFIVTSDD177 pKa = 3.23LVKK180 pKa = 10.48PLILEE185 pKa = 4.23SRR187 pKa = 11.84SQSS190 pKa = 3.15
MM1 pKa = 7.61SNARR5 pKa = 11.84YY6 pKa = 10.52NMISAVSASGCKK18 pKa = 9.02NDD20 pKa = 5.41KK21 pKa = 9.72PFCVHH26 pKa = 6.18KK27 pKa = 10.23VVAVIVIIVSSGITVVNQIGQRR49 pKa = 11.84KK50 pKa = 9.27LSTDD54 pKa = 3.39DD55 pKa = 3.91YY56 pKa = 11.18FVLSLVCDD64 pKa = 5.21AISNIIVVGLSVVGMTASSKK84 pKa = 9.13TKK86 pKa = 10.33QVLTDD91 pKa = 3.42GGEE94 pKa = 3.95NAIEE98 pKa = 4.13LNHH101 pKa = 5.4MPKK104 pKa = 10.3RR105 pKa = 11.84IVKK108 pKa = 10.16KK109 pKa = 10.41LLKK112 pKa = 10.32EE113 pKa = 3.81EE114 pKa = 4.39AKK116 pKa = 10.99NGVDD120 pKa = 4.03FVKK123 pKa = 10.91SIGKK127 pKa = 8.91DD128 pKa = 3.12LKK130 pKa = 11.61SNDD133 pKa = 3.66NKK135 pKa = 10.2PDD137 pKa = 3.49PSAPPKK143 pKa = 10.14YY144 pKa = 10.19EE145 pKa = 4.68KK146 pKa = 9.66IDD148 pKa = 3.41EE149 pKa = 4.27SFEE152 pKa = 4.34LKK154 pKa = 10.62SEE156 pKa = 4.19FAGKK160 pKa = 10.16SLITDD165 pKa = 3.07NRR167 pKa = 11.84TGQSFIVTSDD177 pKa = 3.23LVKK180 pKa = 10.48PLILEE185 pKa = 4.23SRR187 pKa = 11.84SQSS190 pKa = 3.15
Molecular weight: 20.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6305 |
190 |
1223 |
525.4 |
58.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.376 ± 0.457 | 1.427 ± 0.199 |
6.693 ± 0.322 | 5.155 ± 0.27 |
3.759 ± 0.263 | 5.583 ± 0.203 |
2.094 ± 0.228 | 7.407 ± 0.298 |
6.233 ± 0.468 | 9.437 ± 0.373 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.617 ± 0.223 | 6.503 ± 0.508 |
3.236 ± 0.248 | 2.205 ± 0.138 |
5.044 ± 0.46 | 8.374 ± 0.448 |
5.757 ± 0.293 | 7.423 ± 0.533 |
0.508 ± 0.078 | 4.171 ± 0.341 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
