
Donghicola eburneus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Donghicola
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
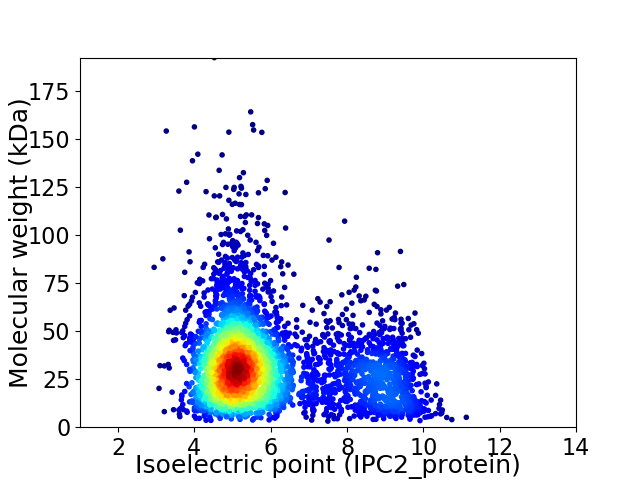
Virtual 2D-PAGE plot for 4047 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
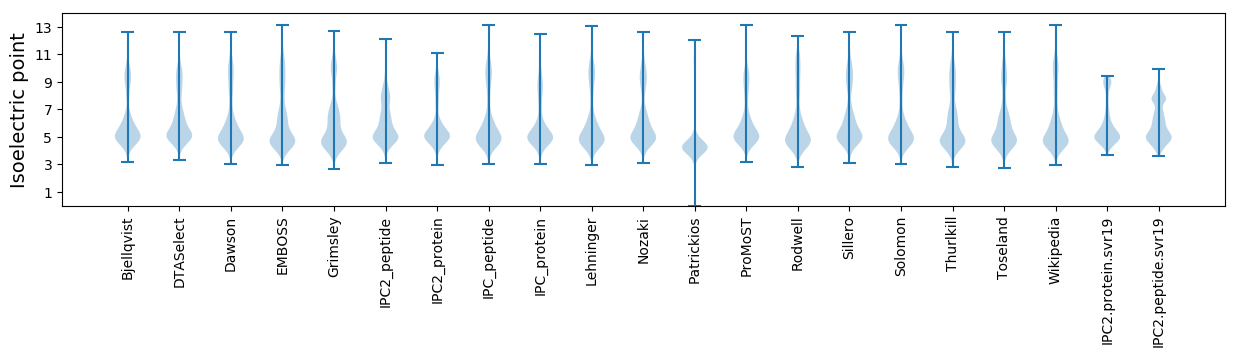
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M4MXU6|A0A1M4MXU6_9RHOB Putative MobA protein OS=Donghicola eburneus OX=393278 GN=mobA1 PE=4 SV=1
MM1 pKa = 7.12STTFQVMYY9 pKa = 10.18LGQLALIDD17 pKa = 4.05SVQGDD22 pKa = 3.89EE23 pKa = 4.13TAEE26 pKa = 3.87NAGAILGSYY35 pKa = 9.75YY36 pKa = 10.9NSTDD40 pKa = 3.08PAYY43 pKa = 10.84DD44 pKa = 3.06HH45 pKa = 7.25TYY47 pKa = 10.96QLTADD52 pKa = 4.57RR53 pKa = 11.84LSEE56 pKa = 4.53DD57 pKa = 4.08ADD59 pKa = 3.56DD60 pKa = 5.34TYY62 pKa = 11.91DD63 pKa = 3.28VDD65 pKa = 4.33NGGGHH70 pKa = 7.32DD71 pKa = 3.46RR72 pKa = 11.84FFVNGTGPYY81 pKa = 10.58DD82 pKa = 3.93FDD84 pKa = 5.07AVAEE88 pKa = 4.06YY89 pKa = 10.57QITLTYY95 pKa = 10.24IDD97 pKa = 4.22GTTANITAYY106 pKa = 10.31VFQDD110 pKa = 3.53TTGHH114 pKa = 5.41TFIAPEE120 pKa = 3.89TTYY123 pKa = 11.35NADD126 pKa = 3.1QAALTDD132 pKa = 4.1KK133 pKa = 10.59PIQSVSLNSVTSYY146 pKa = 10.6TGDD149 pKa = 3.18KK150 pKa = 10.39DD151 pKa = 3.62GDD153 pKa = 4.11MIASRR158 pKa = 11.84VNEE161 pKa = 4.4PYY163 pKa = 10.62AGPVDD168 pKa = 3.87GTNGNDD174 pKa = 3.37SMGVGYY180 pKa = 9.35TDD182 pKa = 3.62AQGHH186 pKa = 7.32AITEE190 pKa = 4.34DD191 pKa = 3.38ADD193 pKa = 3.76VIYY196 pKa = 9.92GYY198 pKa = 10.86DD199 pKa = 3.54GADD202 pKa = 3.58TIDD205 pKa = 3.79GGGGSDD211 pKa = 5.1LIEE214 pKa = 5.04SGAGSDD220 pKa = 3.65VVYY223 pKa = 10.77GGAGNDD229 pKa = 3.54TIYY232 pKa = 11.09YY233 pKa = 7.61GTGNDD238 pKa = 3.87TIYY241 pKa = 11.2GGDD244 pKa = 3.91GNDD247 pKa = 5.62LIDD250 pKa = 5.33DD251 pKa = 4.2EE252 pKa = 6.39DD253 pKa = 4.01GVRR256 pKa = 11.84YY257 pKa = 8.12TGSNEE262 pKa = 3.46IHH264 pKa = 6.41GGEE267 pKa = 4.47GNDD270 pKa = 3.45TVYY273 pKa = 11.01SGDD276 pKa = 3.52GSEE279 pKa = 4.58TIYY282 pKa = 11.37GDD284 pKa = 4.03GGDD287 pKa = 4.43DD288 pKa = 3.52NLFGEE293 pKa = 5.17GGNDD297 pKa = 3.29TLFGGTGNDD306 pKa = 3.74VLWGGSGDD314 pKa = 4.35DD315 pKa = 3.87YY316 pKa = 11.74LAGEE320 pKa = 5.38DD321 pKa = 4.02GNDD324 pKa = 3.38WLVGEE329 pKa = 5.21AGNDD333 pKa = 3.7TLIGGAGNDD342 pKa = 3.5SLYY345 pKa = 11.18GGTGNDD351 pKa = 3.41SMLGGDD357 pKa = 4.34GDD359 pKa = 3.82DD360 pKa = 3.27TFYY363 pKa = 11.58YY364 pKa = 10.42FAGDD368 pKa = 3.74GLDD371 pKa = 3.86TIADD375 pKa = 3.83FGFGNTGSTTDD386 pKa = 4.87GISTNNDD393 pKa = 3.53FIDD396 pKa = 3.88LSSYY400 pKa = 9.61YY401 pKa = 10.78DD402 pKa = 3.28AMNEE406 pKa = 3.68LRR408 pKa = 11.84ADD410 pKa = 3.82FLDD413 pKa = 5.51DD414 pKa = 5.58GILNQSNAADD424 pKa = 3.8YY425 pKa = 11.63SNNTQFQPGDD435 pKa = 3.63GIVFQGATTSDD446 pKa = 3.57FTTDD450 pKa = 2.95STGVVCFTTGTQIRR464 pKa = 11.84TPDD467 pKa = 3.34GDD469 pKa = 3.77RR470 pKa = 11.84PIEE473 pKa = 3.93EE474 pKa = 4.87LKK476 pKa = 11.27VGDD479 pKa = 4.69LVEE482 pKa = 4.44TLDD485 pKa = 5.55GGPQPIKK492 pKa = 10.32WIGTSHH498 pKa = 6.7YY499 pKa = 10.93DD500 pKa = 3.3EE501 pKa = 5.27AALIEE506 pKa = 4.21NPKK509 pKa = 9.99LRR511 pKa = 11.84PIVVSARR518 pKa = 11.84VFGAEE523 pKa = 3.54RR524 pKa = 11.84DD525 pKa = 3.96LVVSRR530 pKa = 11.84QHH532 pKa = 6.91AFLLPKK538 pKa = 10.21EE539 pKa = 4.49DD540 pKa = 3.75LLARR544 pKa = 11.84AIQLVKK550 pKa = 10.44MNVAGVRR557 pKa = 11.84IAHH560 pKa = 5.28GRR562 pKa = 11.84KK563 pKa = 8.72RR564 pKa = 11.84VTYY567 pKa = 8.41HH568 pKa = 6.71HH569 pKa = 6.94IMFEE573 pKa = 3.95KK574 pKa = 10.75HH575 pKa = 4.67EE576 pKa = 4.79LIYY579 pKa = 10.92SEE581 pKa = 5.61GIATEE586 pKa = 4.04SMYY589 pKa = 10.52PGPQALRR596 pKa = 11.84ALCPEE601 pKa = 3.94AMEE604 pKa = 4.28SLLRR608 pKa = 11.84VFPEE612 pKa = 4.18FKK614 pKa = 10.58GIEE617 pKa = 4.07TRR619 pKa = 11.84EE620 pKa = 4.15GAEE623 pKa = 3.65AAYY626 pKa = 10.17GPTARR631 pKa = 11.84EE632 pKa = 4.26MIDD635 pKa = 3.15MSNWRR640 pKa = 11.84AEE642 pKa = 3.89AAAA645 pKa = 4.4
MM1 pKa = 7.12STTFQVMYY9 pKa = 10.18LGQLALIDD17 pKa = 4.05SVQGDD22 pKa = 3.89EE23 pKa = 4.13TAEE26 pKa = 3.87NAGAILGSYY35 pKa = 9.75YY36 pKa = 10.9NSTDD40 pKa = 3.08PAYY43 pKa = 10.84DD44 pKa = 3.06HH45 pKa = 7.25TYY47 pKa = 10.96QLTADD52 pKa = 4.57RR53 pKa = 11.84LSEE56 pKa = 4.53DD57 pKa = 4.08ADD59 pKa = 3.56DD60 pKa = 5.34TYY62 pKa = 11.91DD63 pKa = 3.28VDD65 pKa = 4.33NGGGHH70 pKa = 7.32DD71 pKa = 3.46RR72 pKa = 11.84FFVNGTGPYY81 pKa = 10.58DD82 pKa = 3.93FDD84 pKa = 5.07AVAEE88 pKa = 4.06YY89 pKa = 10.57QITLTYY95 pKa = 10.24IDD97 pKa = 4.22GTTANITAYY106 pKa = 10.31VFQDD110 pKa = 3.53TTGHH114 pKa = 5.41TFIAPEE120 pKa = 3.89TTYY123 pKa = 11.35NADD126 pKa = 3.1QAALTDD132 pKa = 4.1KK133 pKa = 10.59PIQSVSLNSVTSYY146 pKa = 10.6TGDD149 pKa = 3.18KK150 pKa = 10.39DD151 pKa = 3.62GDD153 pKa = 4.11MIASRR158 pKa = 11.84VNEE161 pKa = 4.4PYY163 pKa = 10.62AGPVDD168 pKa = 3.87GTNGNDD174 pKa = 3.37SMGVGYY180 pKa = 9.35TDD182 pKa = 3.62AQGHH186 pKa = 7.32AITEE190 pKa = 4.34DD191 pKa = 3.38ADD193 pKa = 3.76VIYY196 pKa = 9.92GYY198 pKa = 10.86DD199 pKa = 3.54GADD202 pKa = 3.58TIDD205 pKa = 3.79GGGGSDD211 pKa = 5.1LIEE214 pKa = 5.04SGAGSDD220 pKa = 3.65VVYY223 pKa = 10.77GGAGNDD229 pKa = 3.54TIYY232 pKa = 11.09YY233 pKa = 7.61GTGNDD238 pKa = 3.87TIYY241 pKa = 11.2GGDD244 pKa = 3.91GNDD247 pKa = 5.62LIDD250 pKa = 5.33DD251 pKa = 4.2EE252 pKa = 6.39DD253 pKa = 4.01GVRR256 pKa = 11.84YY257 pKa = 8.12TGSNEE262 pKa = 3.46IHH264 pKa = 6.41GGEE267 pKa = 4.47GNDD270 pKa = 3.45TVYY273 pKa = 11.01SGDD276 pKa = 3.52GSEE279 pKa = 4.58TIYY282 pKa = 11.37GDD284 pKa = 4.03GGDD287 pKa = 4.43DD288 pKa = 3.52NLFGEE293 pKa = 5.17GGNDD297 pKa = 3.29TLFGGTGNDD306 pKa = 3.74VLWGGSGDD314 pKa = 4.35DD315 pKa = 3.87YY316 pKa = 11.74LAGEE320 pKa = 5.38DD321 pKa = 4.02GNDD324 pKa = 3.38WLVGEE329 pKa = 5.21AGNDD333 pKa = 3.7TLIGGAGNDD342 pKa = 3.5SLYY345 pKa = 11.18GGTGNDD351 pKa = 3.41SMLGGDD357 pKa = 4.34GDD359 pKa = 3.82DD360 pKa = 3.27TFYY363 pKa = 11.58YY364 pKa = 10.42FAGDD368 pKa = 3.74GLDD371 pKa = 3.86TIADD375 pKa = 3.83FGFGNTGSTTDD386 pKa = 4.87GISTNNDD393 pKa = 3.53FIDD396 pKa = 3.88LSSYY400 pKa = 9.61YY401 pKa = 10.78DD402 pKa = 3.28AMNEE406 pKa = 3.68LRR408 pKa = 11.84ADD410 pKa = 3.82FLDD413 pKa = 5.51DD414 pKa = 5.58GILNQSNAADD424 pKa = 3.8YY425 pKa = 11.63SNNTQFQPGDD435 pKa = 3.63GIVFQGATTSDD446 pKa = 3.57FTTDD450 pKa = 2.95STGVVCFTTGTQIRR464 pKa = 11.84TPDD467 pKa = 3.34GDD469 pKa = 3.77RR470 pKa = 11.84PIEE473 pKa = 3.93EE474 pKa = 4.87LKK476 pKa = 11.27VGDD479 pKa = 4.69LVEE482 pKa = 4.44TLDD485 pKa = 5.55GGPQPIKK492 pKa = 10.32WIGTSHH498 pKa = 6.7YY499 pKa = 10.93DD500 pKa = 3.3EE501 pKa = 5.27AALIEE506 pKa = 4.21NPKK509 pKa = 9.99LRR511 pKa = 11.84PIVVSARR518 pKa = 11.84VFGAEE523 pKa = 3.54RR524 pKa = 11.84DD525 pKa = 3.96LVVSRR530 pKa = 11.84QHH532 pKa = 6.91AFLLPKK538 pKa = 10.21EE539 pKa = 4.49DD540 pKa = 3.75LLARR544 pKa = 11.84AIQLVKK550 pKa = 10.44MNVAGVRR557 pKa = 11.84IAHH560 pKa = 5.28GRR562 pKa = 11.84KK563 pKa = 8.72RR564 pKa = 11.84VTYY567 pKa = 8.41HH568 pKa = 6.71HH569 pKa = 6.94IMFEE573 pKa = 3.95KK574 pKa = 10.75HH575 pKa = 4.67EE576 pKa = 4.79LIYY579 pKa = 10.92SEE581 pKa = 5.61GIATEE586 pKa = 4.04SMYY589 pKa = 10.52PGPQALRR596 pKa = 11.84ALCPEE601 pKa = 3.94AMEE604 pKa = 4.28SLLRR608 pKa = 11.84VFPEE612 pKa = 4.18FKK614 pKa = 10.58GIEE617 pKa = 4.07TRR619 pKa = 11.84EE620 pKa = 4.15GAEE623 pKa = 3.65AAYY626 pKa = 10.17GPTARR631 pKa = 11.84EE632 pKa = 4.26MIDD635 pKa = 3.15MSNWRR640 pKa = 11.84AEE642 pKa = 3.89AAAA645 pKa = 4.4
Molecular weight: 68.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M4N4T5|A0A1M4N4T5_9RHOB Uncharacterized protein OS=Donghicola eburneus OX=393278 GN=KARMA_3320 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1262338 |
30 |
1800 |
311.9 |
33.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.628 ± 0.052 | 0.914 ± 0.013 |
6.166 ± 0.04 | 6.199 ± 0.036 |
3.707 ± 0.024 | 8.455 ± 0.039 |
2.027 ± 0.02 | 5.392 ± 0.025 |
3.521 ± 0.033 | 9.815 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.944 ± 0.015 | 2.843 ± 0.022 |
4.808 ± 0.027 | 3.376 ± 0.023 |
6.211 ± 0.039 | 5.42 ± 0.026 |
5.659 ± 0.028 | 7.193 ± 0.032 |
1.348 ± 0.016 | 2.372 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
