
Microvirga aerophila
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Methylobacteriaceae; Microvirga
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
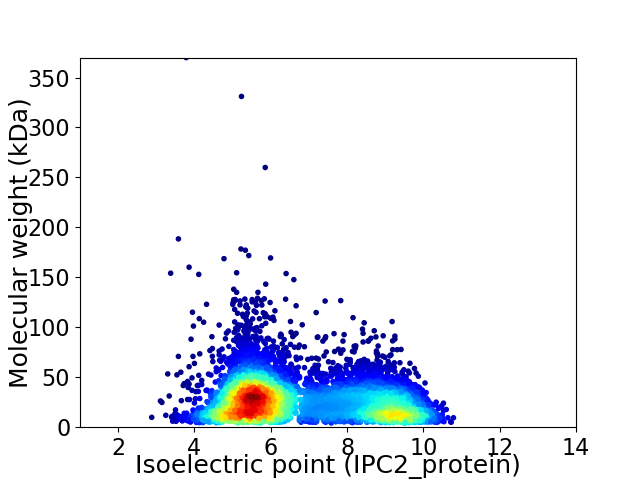
Virtual 2D-PAGE plot for 7161 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A512C525|A0A512C525_9RHIZ IS110 family transposase OS=Microvirga aerophila OX=670291 GN=MAE02_70050 PE=4 SV=1
MM1 pKa = 7.95AILLGNDD8 pKa = 3.21WPNLIRR14 pKa = 11.84GTNASDD20 pKa = 4.12SIWGYY25 pKa = 11.63GDD27 pKa = 5.87DD28 pKa = 4.85DD29 pKa = 4.99DD30 pKa = 6.81LYY32 pKa = 11.25GYY34 pKa = 10.89DD35 pKa = 4.8GDD37 pKa = 4.45DD38 pKa = 4.94DD39 pKa = 5.67IYY41 pKa = 11.38GGWGYY46 pKa = 11.3DD47 pKa = 3.3VLVGGNGDD55 pKa = 4.0DD56 pKa = 3.97YY57 pKa = 11.89LNGGDD62 pKa = 5.35GDD64 pKa = 4.47DD65 pKa = 4.53DD66 pKa = 5.55LYY68 pKa = 11.72GGAAQDD74 pKa = 3.9RR75 pKa = 11.84LIGGAGYY82 pKa = 10.7DD83 pKa = 3.76LLFGQSGNDD92 pKa = 3.51DD93 pKa = 4.53LYY95 pKa = 11.36GQSGYY100 pKa = 11.12DD101 pKa = 3.53DD102 pKa = 5.16LFGGSGLDD110 pKa = 3.59YY111 pKa = 11.31LNGGSSDD118 pKa = 3.66DD119 pKa = 3.77WLDD122 pKa = 3.73GGSGRR127 pKa = 11.84DD128 pKa = 3.31WLVGGTGVDD137 pKa = 2.29IFYY140 pKa = 10.36FKK142 pKa = 10.48RR143 pKa = 11.84GSSGITTASADD154 pKa = 4.06TIADD158 pKa = 3.26WRR160 pKa = 11.84PRR162 pKa = 11.84LDD164 pKa = 3.86WIDD167 pKa = 3.19MSIRR171 pKa = 11.84GTSGNHH177 pKa = 5.77RR178 pKa = 11.84EE179 pKa = 4.18ASTTATSIEE188 pKa = 3.95AAAAQAEE195 pKa = 4.55SRR197 pKa = 11.84FTSASIVHH205 pKa = 5.56VFLYY209 pKa = 10.49NAAADD214 pKa = 3.87RR215 pKa = 11.84GYY217 pKa = 11.39LLSDD221 pKa = 4.13LNNDD225 pKa = 2.91NRR227 pKa = 11.84YY228 pKa = 5.97EE229 pKa = 4.05TGVIILGAGRR239 pKa = 11.84AADD242 pKa = 3.58MDD244 pKa = 4.2YY245 pKa = 11.2RR246 pKa = 11.84WILL249 pKa = 3.35
MM1 pKa = 7.95AILLGNDD8 pKa = 3.21WPNLIRR14 pKa = 11.84GTNASDD20 pKa = 4.12SIWGYY25 pKa = 11.63GDD27 pKa = 5.87DD28 pKa = 4.85DD29 pKa = 4.99DD30 pKa = 6.81LYY32 pKa = 11.25GYY34 pKa = 10.89DD35 pKa = 4.8GDD37 pKa = 4.45DD38 pKa = 4.94DD39 pKa = 5.67IYY41 pKa = 11.38GGWGYY46 pKa = 11.3DD47 pKa = 3.3VLVGGNGDD55 pKa = 4.0DD56 pKa = 3.97YY57 pKa = 11.89LNGGDD62 pKa = 5.35GDD64 pKa = 4.47DD65 pKa = 4.53DD66 pKa = 5.55LYY68 pKa = 11.72GGAAQDD74 pKa = 3.9RR75 pKa = 11.84LIGGAGYY82 pKa = 10.7DD83 pKa = 3.76LLFGQSGNDD92 pKa = 3.51DD93 pKa = 4.53LYY95 pKa = 11.36GQSGYY100 pKa = 11.12DD101 pKa = 3.53DD102 pKa = 5.16LFGGSGLDD110 pKa = 3.59YY111 pKa = 11.31LNGGSSDD118 pKa = 3.66DD119 pKa = 3.77WLDD122 pKa = 3.73GGSGRR127 pKa = 11.84DD128 pKa = 3.31WLVGGTGVDD137 pKa = 2.29IFYY140 pKa = 10.36FKK142 pKa = 10.48RR143 pKa = 11.84GSSGITTASADD154 pKa = 4.06TIADD158 pKa = 3.26WRR160 pKa = 11.84PRR162 pKa = 11.84LDD164 pKa = 3.86WIDD167 pKa = 3.19MSIRR171 pKa = 11.84GTSGNHH177 pKa = 5.77RR178 pKa = 11.84EE179 pKa = 4.18ASTTATSIEE188 pKa = 3.95AAAAQAEE195 pKa = 4.55SRR197 pKa = 11.84FTSASIVHH205 pKa = 5.56VFLYY209 pKa = 10.49NAAADD214 pKa = 3.87RR215 pKa = 11.84GYY217 pKa = 11.39LLSDD221 pKa = 4.13LNNDD225 pKa = 2.91NRR227 pKa = 11.84YY228 pKa = 5.97EE229 pKa = 4.05TGVIILGAGRR239 pKa = 11.84AADD242 pKa = 3.58MDD244 pKa = 4.2YY245 pKa = 11.2RR246 pKa = 11.84WILL249 pKa = 3.35
Molecular weight: 26.57 kDa
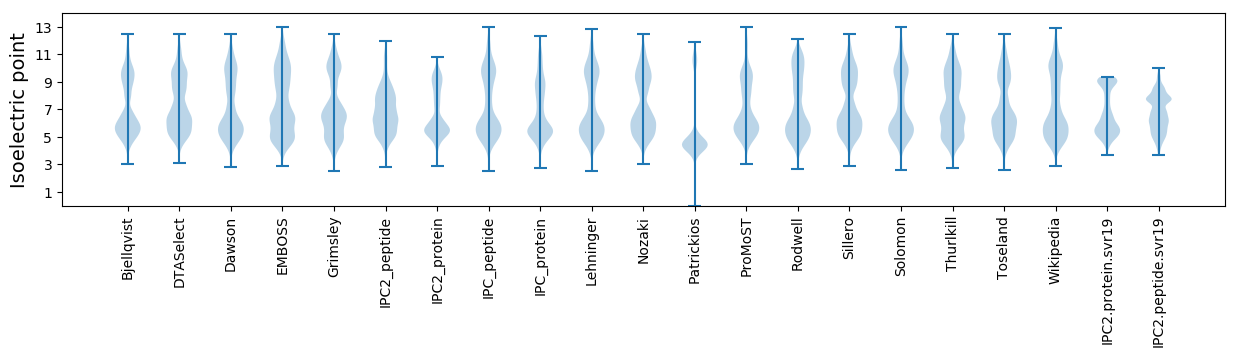
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A512BW53|A0A512BW53_9RHIZ Uncharacterized protein OS=Microvirga aerophila OX=670291 GN=MAE02_38510 PE=4 SV=1
MM1 pKa = 7.13EE2 pKa = 4.1VSVRR6 pKa = 11.84EE7 pKa = 4.05VLSGITTMRR16 pKa = 11.84DD17 pKa = 2.96MVKK20 pKa = 10.52AFQNEE25 pKa = 3.96AHH27 pKa = 6.79CRR29 pKa = 11.84RR30 pKa = 11.84LVQAMVWPKK39 pKa = 10.88GRR41 pKa = 11.84ICPACGYY48 pKa = 9.46RR49 pKa = 11.84RR50 pKa = 11.84SIALTGRR57 pKa = 11.84EE58 pKa = 3.66NGKK61 pKa = 9.37RR62 pKa = 11.84ARR64 pKa = 11.84SGLYY68 pKa = 8.38QCSSGTCRR76 pKa = 11.84FQFTVTTRR84 pKa = 11.84TPLHH88 pKa = 5.73ATKK91 pKa = 10.73LPLRR95 pKa = 11.84TWLSGLWLMLQSDD108 pKa = 4.34KK109 pKa = 11.27GISSIRR115 pKa = 11.84LAEE118 pKa = 4.14ALGVSQPTAWRR129 pKa = 11.84MGHH132 pKa = 6.73ALRR135 pKa = 11.84LMVGRR140 pKa = 11.84EE141 pKa = 3.55RR142 pKa = 11.84ALDD145 pKa = 3.65GVVEE149 pKa = 4.01IDD151 pKa = 3.35GLYY154 pKa = 10.73LGGKK158 pKa = 7.69PRR160 pKa = 11.84RR161 pKa = 11.84EE162 pKa = 3.7VDD164 pKa = 3.22YY165 pKa = 10.97SPPGRR170 pKa = 11.84GRR172 pKa = 11.84KK173 pKa = 8.15GQPNDD178 pKa = 3.26LHH180 pKa = 6.71EE181 pKa = 5.36GGVTT185 pKa = 3.28
MM1 pKa = 7.13EE2 pKa = 4.1VSVRR6 pKa = 11.84EE7 pKa = 4.05VLSGITTMRR16 pKa = 11.84DD17 pKa = 2.96MVKK20 pKa = 10.52AFQNEE25 pKa = 3.96AHH27 pKa = 6.79CRR29 pKa = 11.84RR30 pKa = 11.84LVQAMVWPKK39 pKa = 10.88GRR41 pKa = 11.84ICPACGYY48 pKa = 9.46RR49 pKa = 11.84RR50 pKa = 11.84SIALTGRR57 pKa = 11.84EE58 pKa = 3.66NGKK61 pKa = 9.37RR62 pKa = 11.84ARR64 pKa = 11.84SGLYY68 pKa = 8.38QCSSGTCRR76 pKa = 11.84FQFTVTTRR84 pKa = 11.84TPLHH88 pKa = 5.73ATKK91 pKa = 10.73LPLRR95 pKa = 11.84TWLSGLWLMLQSDD108 pKa = 4.34KK109 pKa = 11.27GISSIRR115 pKa = 11.84LAEE118 pKa = 4.14ALGVSQPTAWRR129 pKa = 11.84MGHH132 pKa = 6.73ALRR135 pKa = 11.84LMVGRR140 pKa = 11.84EE141 pKa = 3.55RR142 pKa = 11.84ALDD145 pKa = 3.65GVVEE149 pKa = 4.01IDD151 pKa = 3.35GLYY154 pKa = 10.73LGGKK158 pKa = 7.69PRR160 pKa = 11.84RR161 pKa = 11.84EE162 pKa = 3.7VDD164 pKa = 3.22YY165 pKa = 10.97SPPGRR170 pKa = 11.84GRR172 pKa = 11.84KK173 pKa = 8.15GQPNDD178 pKa = 3.26LHH180 pKa = 6.71EE181 pKa = 5.36GGVTT185 pKa = 3.28
Molecular weight: 20.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1921182 |
39 |
3586 |
268.3 |
29.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.74 ± 0.035 | 0.815 ± 0.009 |
5.523 ± 0.028 | 5.864 ± 0.03 |
3.649 ± 0.02 | 8.397 ± 0.035 |
2.113 ± 0.015 | 5.173 ± 0.021 |
3.273 ± 0.027 | 10.193 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.366 ± 0.014 | 2.616 ± 0.017 |
5.316 ± 0.02 | 3.376 ± 0.019 |
7.406 ± 0.037 | 5.648 ± 0.022 |
5.368 ± 0.025 | 7.528 ± 0.023 |
1.385 ± 0.012 | 2.251 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
