
Hubei tombus-like virus 31
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.02
Get precalculated fractions of proteins
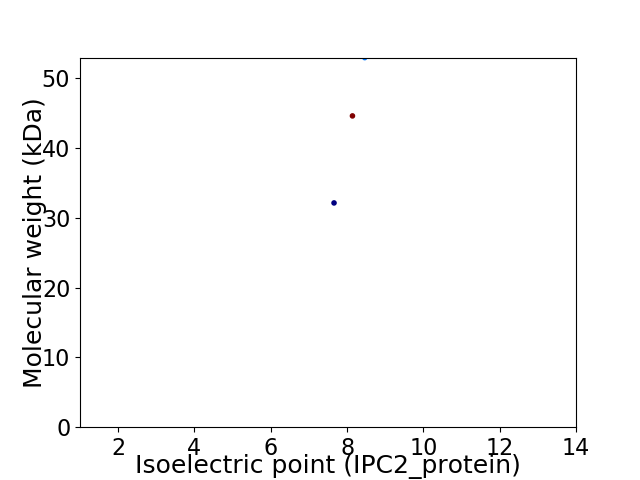
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
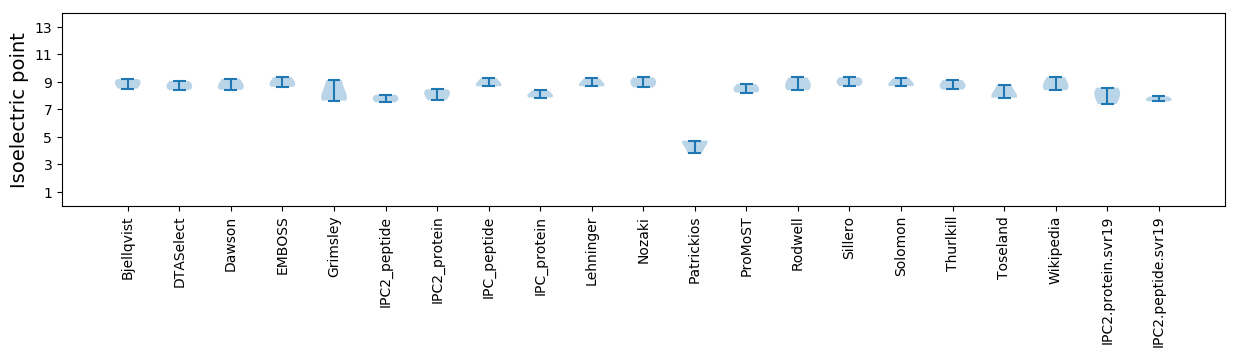
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGW3|A0A1L3KGW3_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 31 OX=1923279 PE=4 SV=1
MM1 pKa = 6.81NTNTTTHH8 pKa = 5.81EE9 pKa = 4.18RR10 pKa = 11.84TLKK13 pKa = 10.65SGIVVKK19 pKa = 7.56FTRR22 pKa = 11.84SPGTLLSKK30 pKa = 10.11DD31 pKa = 3.55TEE33 pKa = 4.57TEE35 pKa = 4.0GTIVTDD41 pKa = 3.66GWEE44 pKa = 4.09SSSRR48 pKa = 11.84SQTVSEE54 pKa = 4.3PTNLAAALEE63 pKa = 4.22PVLTKK68 pKa = 10.69GVVVASVVRR77 pKa = 11.84PQRR80 pKa = 11.84QAPEE84 pKa = 4.45SPVLTLRR91 pKa = 11.84AIRR94 pKa = 11.84RR95 pKa = 11.84GTRR98 pKa = 11.84AGRR101 pKa = 11.84SVRR104 pKa = 11.84RR105 pKa = 11.84SAVFKK110 pKa = 10.42EE111 pKa = 4.1LQDD114 pKa = 4.22YY115 pKa = 8.39PLSWAVIEE123 pKa = 4.05QLRR126 pKa = 11.84ILATEE131 pKa = 3.83ICLGKK136 pKa = 10.03EE137 pKa = 3.65LNVEE141 pKa = 3.79NFRR144 pKa = 11.84IFVRR148 pKa = 11.84KK149 pKa = 9.86FEE151 pKa = 5.07DD152 pKa = 3.51KK153 pKa = 10.32LTLTEE158 pKa = 4.16VYY160 pKa = 10.28YY161 pKa = 10.7SQSRR165 pKa = 11.84LDD167 pKa = 3.69SVEE170 pKa = 4.91DD171 pKa = 3.76ILWDD175 pKa = 3.61EE176 pKa = 4.71APAHH180 pKa = 5.75AVAVIKK186 pKa = 10.8ISDD189 pKa = 3.97DD190 pKa = 3.37RR191 pKa = 11.84LSATRR196 pKa = 11.84SFRR199 pKa = 11.84LGSALEE205 pKa = 4.13ASLPAAAYY213 pKa = 10.2RR214 pKa = 11.84DD215 pKa = 3.88VANTASAVDD224 pKa = 3.42DD225 pKa = 4.1MFRR228 pKa = 11.84EE229 pKa = 4.89LYY231 pKa = 10.43LLHH234 pKa = 6.56NPGVIEE240 pKa = 4.88RR241 pKa = 11.84RR242 pKa = 11.84NIQQGRR248 pKa = 11.84IEE250 pKa = 4.36YY251 pKa = 10.73GSDD254 pKa = 2.41SWLWRR259 pKa = 11.84GLEE262 pKa = 3.96SVYY265 pKa = 10.82YY266 pKa = 10.59GSAARR271 pKa = 11.84WNWLVGKK278 pKa = 9.72QLAVPTAPQCC288 pKa = 3.99
MM1 pKa = 6.81NTNTTTHH8 pKa = 5.81EE9 pKa = 4.18RR10 pKa = 11.84TLKK13 pKa = 10.65SGIVVKK19 pKa = 7.56FTRR22 pKa = 11.84SPGTLLSKK30 pKa = 10.11DD31 pKa = 3.55TEE33 pKa = 4.57TEE35 pKa = 4.0GTIVTDD41 pKa = 3.66GWEE44 pKa = 4.09SSSRR48 pKa = 11.84SQTVSEE54 pKa = 4.3PTNLAAALEE63 pKa = 4.22PVLTKK68 pKa = 10.69GVVVASVVRR77 pKa = 11.84PQRR80 pKa = 11.84QAPEE84 pKa = 4.45SPVLTLRR91 pKa = 11.84AIRR94 pKa = 11.84RR95 pKa = 11.84GTRR98 pKa = 11.84AGRR101 pKa = 11.84SVRR104 pKa = 11.84RR105 pKa = 11.84SAVFKK110 pKa = 10.42EE111 pKa = 4.1LQDD114 pKa = 4.22YY115 pKa = 8.39PLSWAVIEE123 pKa = 4.05QLRR126 pKa = 11.84ILATEE131 pKa = 3.83ICLGKK136 pKa = 10.03EE137 pKa = 3.65LNVEE141 pKa = 3.79NFRR144 pKa = 11.84IFVRR148 pKa = 11.84KK149 pKa = 9.86FEE151 pKa = 5.07DD152 pKa = 3.51KK153 pKa = 10.32LTLTEE158 pKa = 4.16VYY160 pKa = 10.28YY161 pKa = 10.7SQSRR165 pKa = 11.84LDD167 pKa = 3.69SVEE170 pKa = 4.91DD171 pKa = 3.76ILWDD175 pKa = 3.61EE176 pKa = 4.71APAHH180 pKa = 5.75AVAVIKK186 pKa = 10.8ISDD189 pKa = 3.97DD190 pKa = 3.37RR191 pKa = 11.84LSATRR196 pKa = 11.84SFRR199 pKa = 11.84LGSALEE205 pKa = 4.13ASLPAAAYY213 pKa = 10.2RR214 pKa = 11.84DD215 pKa = 3.88VANTASAVDD224 pKa = 3.42DD225 pKa = 4.1MFRR228 pKa = 11.84EE229 pKa = 4.89LYY231 pKa = 10.43LLHH234 pKa = 6.56NPGVIEE240 pKa = 4.88RR241 pKa = 11.84RR242 pKa = 11.84NIQQGRR248 pKa = 11.84IEE250 pKa = 4.36YY251 pKa = 10.73GSDD254 pKa = 2.41SWLWRR259 pKa = 11.84GLEE262 pKa = 3.96SVYY265 pKa = 10.82YY266 pKa = 10.59GSAARR271 pKa = 11.84WNWLVGKK278 pKa = 9.72QLAVPTAPQCC288 pKa = 3.99
Molecular weight: 32.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGW3|A0A1L3KGW3_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 31 OX=1923279 PE=4 SV=1
MM1 pKa = 7.46NARR4 pKa = 11.84LALSQRR10 pKa = 11.84LEE12 pKa = 3.93IPANVALRR20 pKa = 11.84KK21 pKa = 10.0VEE23 pKa = 3.86INKK26 pKa = 9.59DD27 pKa = 3.08HH28 pKa = 5.41THH30 pKa = 5.26WVEE33 pKa = 3.68LYY35 pKa = 10.73KK36 pKa = 10.32IVSKK40 pKa = 10.5LARR43 pKa = 11.84AVSHH47 pKa = 6.54IRR49 pKa = 11.84PLTPAEE55 pKa = 3.98FLKK58 pKa = 10.17TANSRR63 pKa = 11.84QRR65 pKa = 11.84PVWEE69 pKa = 3.57RR70 pKa = 11.84ALRR73 pKa = 11.84QDD75 pKa = 3.5RR76 pKa = 11.84RR77 pKa = 11.84FRR79 pKa = 11.84PSWMTYY85 pKa = 10.36DD86 pKa = 3.52GFVKK90 pKa = 9.93IEE92 pKa = 3.9KK93 pKa = 9.24YY94 pKa = 10.52SYY96 pKa = 7.92YY97 pKa = 10.77EE98 pKa = 3.65KK99 pKa = 10.32MNRR102 pKa = 11.84IPRR105 pKa = 11.84LILPPSDD112 pKa = 3.19HH113 pKa = 6.98AKK115 pKa = 10.38VVMGMHH121 pKa = 6.88IKK123 pKa = 10.31PIEE126 pKa = 3.92KK127 pKa = 9.77HH128 pKa = 5.43LKK130 pKa = 9.3EE131 pKa = 4.24VVGPGNVYY139 pKa = 10.42PFMAKK144 pKa = 10.02GMSSRR149 pKa = 11.84QLAEE153 pKa = 3.42RR154 pKa = 11.84FMRR157 pKa = 11.84MAEE160 pKa = 3.96QFKK163 pKa = 10.35RR164 pKa = 11.84PTFISIDD171 pKa = 3.52MSKK174 pKa = 10.55CDD176 pKa = 3.52STIGPRR182 pKa = 11.84LKK184 pKa = 10.51RR185 pKa = 11.84LEE187 pKa = 3.95NMVFTNYY194 pKa = 9.74HH195 pKa = 5.67SSSDD199 pKa = 3.02HH200 pKa = 5.46RR201 pKa = 11.84RR202 pKa = 11.84CMKK205 pKa = 10.45EE206 pKa = 3.75GEE208 pKa = 4.22KK209 pKa = 10.18TFMKK213 pKa = 10.35VRR215 pKa = 11.84LHH217 pKa = 6.72CNDD220 pKa = 2.78GTTEE224 pKa = 3.87TSEE227 pKa = 4.1IPQCRR232 pKa = 11.84ASGTAHH238 pKa = 6.53TGAGNTVLVYY248 pKa = 10.24GASAVVLRR256 pKa = 11.84GINNEE261 pKa = 3.95VFSNGDD267 pKa = 3.25DD268 pKa = 3.64TILIVEE274 pKa = 4.72ASDD277 pKa = 3.27SAEE280 pKa = 3.93VIRR283 pKa = 11.84RR284 pKa = 11.84INDD287 pKa = 3.05HH288 pKa = 6.73EE289 pKa = 4.43YY290 pKa = 10.72SVFGFDD296 pKa = 4.35VRR298 pKa = 11.84IEE300 pKa = 4.0QVATDD305 pKa = 3.53IEE307 pKa = 4.64DD308 pKa = 4.01VFWCQCYY315 pKa = 8.25YY316 pKa = 9.84TVRR319 pKa = 11.84KK320 pKa = 10.04DD321 pKa = 3.38GPIWIRR327 pKa = 11.84DD328 pKa = 3.59YY329 pKa = 11.43RR330 pKa = 11.84KK331 pKa = 9.69VLQTILSNEE340 pKa = 4.31NYY342 pKa = 10.34GSPNWLSYY350 pKa = 11.17LSTICLGEE358 pKa = 4.23GSTNPGQPIVAPLVSTILKK377 pKa = 9.66LRR379 pKa = 11.84HH380 pKa = 5.51KK381 pKa = 10.57RR382 pKa = 11.84MRR384 pKa = 11.84LPNEE388 pKa = 3.81NQTTRR393 pKa = 11.84RR394 pKa = 11.84WEE396 pKa = 3.99LEE398 pKa = 3.68GCPSVEE404 pKa = 3.99EE405 pKa = 4.37LDD407 pKa = 5.04LEE409 pKa = 4.64VTAQDD414 pKa = 3.51RR415 pKa = 11.84ACFHH419 pKa = 6.3ARR421 pKa = 11.84YY422 pKa = 9.42GISPVEE428 pKa = 4.11QLRR431 pKa = 11.84MEE433 pKa = 4.52AVNNLALRR441 pKa = 11.84GLGDD445 pKa = 3.42GPVRR449 pKa = 11.84HH450 pKa = 6.55EE451 pKa = 3.62YY452 pKa = 7.52WQNRR456 pKa = 11.84IHH458 pKa = 7.02PVGG461 pKa = 3.38
MM1 pKa = 7.46NARR4 pKa = 11.84LALSQRR10 pKa = 11.84LEE12 pKa = 3.93IPANVALRR20 pKa = 11.84KK21 pKa = 10.0VEE23 pKa = 3.86INKK26 pKa = 9.59DD27 pKa = 3.08HH28 pKa = 5.41THH30 pKa = 5.26WVEE33 pKa = 3.68LYY35 pKa = 10.73KK36 pKa = 10.32IVSKK40 pKa = 10.5LARR43 pKa = 11.84AVSHH47 pKa = 6.54IRR49 pKa = 11.84PLTPAEE55 pKa = 3.98FLKK58 pKa = 10.17TANSRR63 pKa = 11.84QRR65 pKa = 11.84PVWEE69 pKa = 3.57RR70 pKa = 11.84ALRR73 pKa = 11.84QDD75 pKa = 3.5RR76 pKa = 11.84RR77 pKa = 11.84FRR79 pKa = 11.84PSWMTYY85 pKa = 10.36DD86 pKa = 3.52GFVKK90 pKa = 9.93IEE92 pKa = 3.9KK93 pKa = 9.24YY94 pKa = 10.52SYY96 pKa = 7.92YY97 pKa = 10.77EE98 pKa = 3.65KK99 pKa = 10.32MNRR102 pKa = 11.84IPRR105 pKa = 11.84LILPPSDD112 pKa = 3.19HH113 pKa = 6.98AKK115 pKa = 10.38VVMGMHH121 pKa = 6.88IKK123 pKa = 10.31PIEE126 pKa = 3.92KK127 pKa = 9.77HH128 pKa = 5.43LKK130 pKa = 9.3EE131 pKa = 4.24VVGPGNVYY139 pKa = 10.42PFMAKK144 pKa = 10.02GMSSRR149 pKa = 11.84QLAEE153 pKa = 3.42RR154 pKa = 11.84FMRR157 pKa = 11.84MAEE160 pKa = 3.96QFKK163 pKa = 10.35RR164 pKa = 11.84PTFISIDD171 pKa = 3.52MSKK174 pKa = 10.55CDD176 pKa = 3.52STIGPRR182 pKa = 11.84LKK184 pKa = 10.51RR185 pKa = 11.84LEE187 pKa = 3.95NMVFTNYY194 pKa = 9.74HH195 pKa = 5.67SSSDD199 pKa = 3.02HH200 pKa = 5.46RR201 pKa = 11.84RR202 pKa = 11.84CMKK205 pKa = 10.45EE206 pKa = 3.75GEE208 pKa = 4.22KK209 pKa = 10.18TFMKK213 pKa = 10.35VRR215 pKa = 11.84LHH217 pKa = 6.72CNDD220 pKa = 2.78GTTEE224 pKa = 3.87TSEE227 pKa = 4.1IPQCRR232 pKa = 11.84ASGTAHH238 pKa = 6.53TGAGNTVLVYY248 pKa = 10.24GASAVVLRR256 pKa = 11.84GINNEE261 pKa = 3.95VFSNGDD267 pKa = 3.25DD268 pKa = 3.64TILIVEE274 pKa = 4.72ASDD277 pKa = 3.27SAEE280 pKa = 3.93VIRR283 pKa = 11.84RR284 pKa = 11.84INDD287 pKa = 3.05HH288 pKa = 6.73EE289 pKa = 4.43YY290 pKa = 10.72SVFGFDD296 pKa = 4.35VRR298 pKa = 11.84IEE300 pKa = 4.0QVATDD305 pKa = 3.53IEE307 pKa = 4.64DD308 pKa = 4.01VFWCQCYY315 pKa = 8.25YY316 pKa = 9.84TVRR319 pKa = 11.84KK320 pKa = 10.04DD321 pKa = 3.38GPIWIRR327 pKa = 11.84DD328 pKa = 3.59YY329 pKa = 11.43RR330 pKa = 11.84KK331 pKa = 9.69VLQTILSNEE340 pKa = 4.31NYY342 pKa = 10.34GSPNWLSYY350 pKa = 11.17LSTICLGEE358 pKa = 4.23GSTNPGQPIVAPLVSTILKK377 pKa = 9.66LRR379 pKa = 11.84HH380 pKa = 5.51KK381 pKa = 10.57RR382 pKa = 11.84MRR384 pKa = 11.84LPNEE388 pKa = 3.81NQTTRR393 pKa = 11.84RR394 pKa = 11.84WEE396 pKa = 3.99LEE398 pKa = 3.68GCPSVEE404 pKa = 3.99EE405 pKa = 4.37LDD407 pKa = 5.04LEE409 pKa = 4.64VTAQDD414 pKa = 3.51RR415 pKa = 11.84ACFHH419 pKa = 6.3ARR421 pKa = 11.84YY422 pKa = 9.42GISPVEE428 pKa = 4.11QLRR431 pKa = 11.84MEE433 pKa = 4.52AVNNLALRR441 pKa = 11.84GLGDD445 pKa = 3.42GPVRR449 pKa = 11.84HH450 pKa = 6.55EE451 pKa = 3.62YY452 pKa = 7.52WQNRR456 pKa = 11.84IHH458 pKa = 7.02PVGG461 pKa = 3.38
Molecular weight: 52.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1165 |
288 |
461 |
388.3 |
43.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.124 ± 0.805 | 1.545 ± 0.296 |
4.12 ± 0.24 | 5.579 ± 1.34 |
3.777 ± 0.819 | 7.039 ± 1.294 |
1.974 ± 0.595 | 5.322 ± 0.368 |
4.549 ± 0.37 | 7.639 ± 0.958 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.403 ± 0.597 | 4.807 ± 0.628 |
4.549 ± 0.307 | 3.262 ± 0.107 |
6.953 ± 1.744 | 8.412 ± 0.929 |
7.639 ± 1.317 | 8.155 ± 0.325 |
1.459 ± 0.482 | 3.691 ± 0.438 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
