
Ciceribacter sp. F8825
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Ciceribacter; unclassified Ciceribacter
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
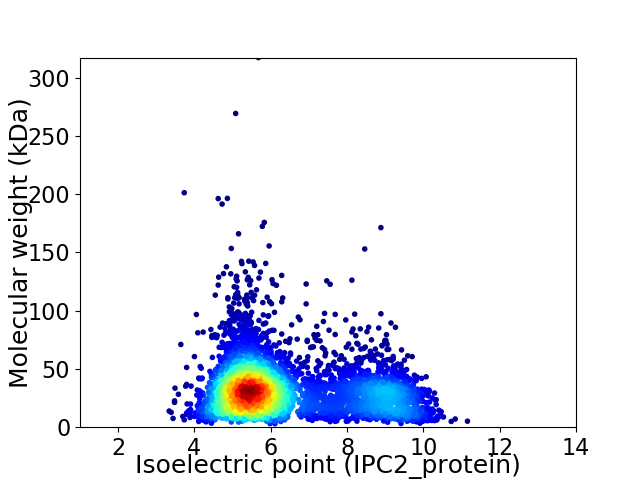
Virtual 2D-PAGE plot for 4841 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q2T1I1|A0A4Q2T1I1_9RHIZ Uncharacterized protein OS=Ciceribacter sp. F8825 OX=2509717 GN=EUU22_12290 PE=4 SV=1
MM1 pKa = 7.26ARR3 pKa = 11.84SVEE6 pKa = 4.11NALGIPLYY14 pKa = 11.03YY15 pKa = 10.51SGTSTAWFSATGSGPSLYY33 pKa = 9.26GTAGNDD39 pKa = 4.12SIWGDD44 pKa = 3.38SSVNVTMYY52 pKa = 10.91GGTGDD57 pKa = 4.88DD58 pKa = 3.2IYY60 pKa = 11.69YY61 pKa = 10.45LYY63 pKa = 10.06STINRR68 pKa = 11.84AVEE71 pKa = 3.96APGEE75 pKa = 4.44GIDD78 pKa = 5.03TIKK81 pKa = 9.65TWMSYY86 pKa = 7.28TLPEE90 pKa = 4.05NFEE93 pKa = 4.33NLSVTGDD100 pKa = 3.24VRR102 pKa = 11.84HH103 pKa = 6.52AFGNDD108 pKa = 2.87LDD110 pKa = 4.91NIISGSSNRR119 pKa = 11.84QTFDD123 pKa = 2.73GGAGNDD129 pKa = 3.49VLTGGGGADD138 pKa = 3.24IFIVDD143 pKa = 4.52KK144 pKa = 11.6GNGSDD149 pKa = 5.15LITDD153 pKa = 4.27FRR155 pKa = 11.84ADD157 pKa = 3.28DD158 pKa = 4.18TIRR161 pKa = 11.84LEE163 pKa = 4.9GYY165 pKa = 10.25AFTSFAEE172 pKa = 4.59VSSHH176 pKa = 5.65LTQEE180 pKa = 4.34GTDD183 pKa = 3.36VRR185 pKa = 11.84LDD187 pKa = 3.69LGDD190 pKa = 4.2GEE192 pKa = 4.99SLVFADD198 pKa = 4.62VAVGDD203 pKa = 4.42LGANQFRR210 pKa = 11.84LSLDD214 pKa = 3.47RR215 pKa = 11.84SHH217 pKa = 7.07LTQTFSDD224 pKa = 4.18DD225 pKa = 4.16FNTLSLNDD233 pKa = 4.02GSGGTWDD240 pKa = 4.2PKK242 pKa = 10.55FWWAPEE248 pKa = 4.16KK249 pKa = 10.86GSSLAGNGEE258 pKa = 4.33LQWYY262 pKa = 9.12INPAYY267 pKa = 10.03EE268 pKa = 4.45GTSEE272 pKa = 4.11VNPFSVEE279 pKa = 3.96NGVLTITAEE288 pKa = 3.89QAPPSIQSEE297 pKa = 4.36IEE299 pKa = 4.37GYY301 pKa = 10.67DD302 pKa = 3.46YY303 pKa = 11.2TSGMLTTYY311 pKa = 10.96SSFAQTYY318 pKa = 8.7GYY320 pKa = 10.77FEE322 pKa = 4.72IRR324 pKa = 11.84ADD326 pKa = 3.68MPDD329 pKa = 3.52EE330 pKa = 4.25QGTWPAFWLLPADD343 pKa = 4.39GSWPPEE349 pKa = 3.76LDD351 pKa = 3.52VVEE354 pKa = 5.06MRR356 pKa = 11.84GQDD359 pKa = 3.54PNTVHH364 pKa = 6.68VSVHH368 pKa = 5.61SNEE371 pKa = 3.98TGQRR375 pKa = 11.84TTTSTAVQVSGTEE388 pKa = 4.0GFHH391 pKa = 6.78NYY393 pKa = 8.39GVLWTEE399 pKa = 4.54EE400 pKa = 4.13EE401 pKa = 4.36IVWYY405 pKa = 10.18FDD407 pKa = 3.44DD408 pKa = 4.07VAVARR413 pKa = 11.84ADD415 pKa = 3.96TPADD419 pKa = 3.41MHH421 pKa = 7.3DD422 pKa = 3.79PMYY425 pKa = 10.22MIVNLAVGGSAGTPADD441 pKa = 3.63GLAGGAEE448 pKa = 3.94MQIDD452 pKa = 4.22YY453 pKa = 10.83IKK455 pKa = 10.56AYY457 pKa = 10.71SLDD460 pKa = 3.73DD461 pKa = 3.75VMPAQQQQTSEE472 pKa = 3.9WLLL475 pKa = 3.6
MM1 pKa = 7.26ARR3 pKa = 11.84SVEE6 pKa = 4.11NALGIPLYY14 pKa = 11.03YY15 pKa = 10.51SGTSTAWFSATGSGPSLYY33 pKa = 9.26GTAGNDD39 pKa = 4.12SIWGDD44 pKa = 3.38SSVNVTMYY52 pKa = 10.91GGTGDD57 pKa = 4.88DD58 pKa = 3.2IYY60 pKa = 11.69YY61 pKa = 10.45LYY63 pKa = 10.06STINRR68 pKa = 11.84AVEE71 pKa = 3.96APGEE75 pKa = 4.44GIDD78 pKa = 5.03TIKK81 pKa = 9.65TWMSYY86 pKa = 7.28TLPEE90 pKa = 4.05NFEE93 pKa = 4.33NLSVTGDD100 pKa = 3.24VRR102 pKa = 11.84HH103 pKa = 6.52AFGNDD108 pKa = 2.87LDD110 pKa = 4.91NIISGSSNRR119 pKa = 11.84QTFDD123 pKa = 2.73GGAGNDD129 pKa = 3.49VLTGGGGADD138 pKa = 3.24IFIVDD143 pKa = 4.52KK144 pKa = 11.6GNGSDD149 pKa = 5.15LITDD153 pKa = 4.27FRR155 pKa = 11.84ADD157 pKa = 3.28DD158 pKa = 4.18TIRR161 pKa = 11.84LEE163 pKa = 4.9GYY165 pKa = 10.25AFTSFAEE172 pKa = 4.59VSSHH176 pKa = 5.65LTQEE180 pKa = 4.34GTDD183 pKa = 3.36VRR185 pKa = 11.84LDD187 pKa = 3.69LGDD190 pKa = 4.2GEE192 pKa = 4.99SLVFADD198 pKa = 4.62VAVGDD203 pKa = 4.42LGANQFRR210 pKa = 11.84LSLDD214 pKa = 3.47RR215 pKa = 11.84SHH217 pKa = 7.07LTQTFSDD224 pKa = 4.18DD225 pKa = 4.16FNTLSLNDD233 pKa = 4.02GSGGTWDD240 pKa = 4.2PKK242 pKa = 10.55FWWAPEE248 pKa = 4.16KK249 pKa = 10.86GSSLAGNGEE258 pKa = 4.33LQWYY262 pKa = 9.12INPAYY267 pKa = 10.03EE268 pKa = 4.45GTSEE272 pKa = 4.11VNPFSVEE279 pKa = 3.96NGVLTITAEE288 pKa = 3.89QAPPSIQSEE297 pKa = 4.36IEE299 pKa = 4.37GYY301 pKa = 10.67DD302 pKa = 3.46YY303 pKa = 11.2TSGMLTTYY311 pKa = 10.96SSFAQTYY318 pKa = 8.7GYY320 pKa = 10.77FEE322 pKa = 4.72IRR324 pKa = 11.84ADD326 pKa = 3.68MPDD329 pKa = 3.52EE330 pKa = 4.25QGTWPAFWLLPADD343 pKa = 4.39GSWPPEE349 pKa = 3.76LDD351 pKa = 3.52VVEE354 pKa = 5.06MRR356 pKa = 11.84GQDD359 pKa = 3.54PNTVHH364 pKa = 6.68VSVHH368 pKa = 5.61SNEE371 pKa = 3.98TGQRR375 pKa = 11.84TTTSTAVQVSGTEE388 pKa = 4.0GFHH391 pKa = 6.78NYY393 pKa = 8.39GVLWTEE399 pKa = 4.54EE400 pKa = 4.13EE401 pKa = 4.36IVWYY405 pKa = 10.18FDD407 pKa = 3.44DD408 pKa = 4.07VAVARR413 pKa = 11.84ADD415 pKa = 3.96TPADD419 pKa = 3.41MHH421 pKa = 7.3DD422 pKa = 3.79PMYY425 pKa = 10.22MIVNLAVGGSAGTPADD441 pKa = 3.63GLAGGAEE448 pKa = 3.94MQIDD452 pKa = 4.22YY453 pKa = 10.83IKK455 pKa = 10.56AYY457 pKa = 10.71SLDD460 pKa = 3.73DD461 pKa = 3.75VMPAQQQQTSEE472 pKa = 3.9WLLL475 pKa = 3.6
Molecular weight: 51.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1RPQ3|A0A4V1RPQ3_9RHIZ Type II toxin-antitoxin system HicB family antitoxin OS=Ciceribacter sp. F8825 OX=2509717 GN=EUU22_19010 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATNGGRR28 pKa = 11.84KK29 pKa = 8.93VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATNGGRR28 pKa = 11.84KK29 pKa = 8.93VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1479523 |
27 |
2833 |
305.6 |
33.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.12 ± 0.053 | 0.837 ± 0.012 |
5.663 ± 0.028 | 6.078 ± 0.033 |
3.892 ± 0.024 | 8.485 ± 0.036 |
1.998 ± 0.018 | 5.37 ± 0.025 |
3.483 ± 0.03 | 9.996 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.55 ± 0.015 | 2.624 ± 0.021 |
4.957 ± 0.024 | 2.857 ± 0.022 |
7.022 ± 0.036 | 5.537 ± 0.026 |
5.337 ± 0.024 | 7.628 ± 0.026 |
1.268 ± 0.014 | 2.299 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
