
Rhodoblastus sphagnicola
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Beijerinckiaceae; Rhodoblastus
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
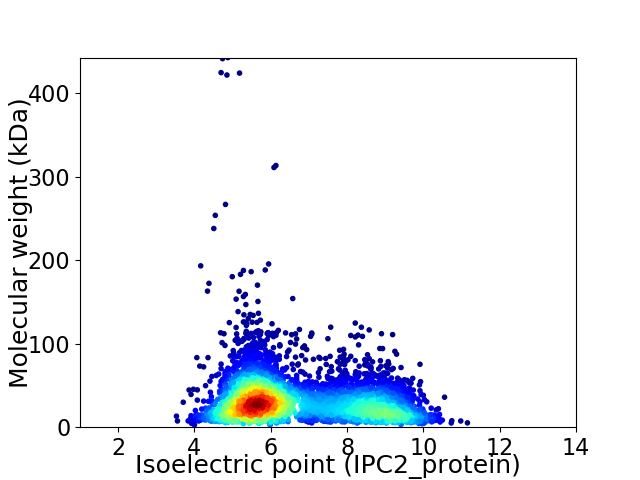
Virtual 2D-PAGE plot for 4616 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S6NAX3|A0A2S6NAX3_9RHIZ Parvulin-like PPIase OS=Rhodoblastus sphagnicola OX=333368 GN=CCR94_08145 PE=3 SV=1
MM1 pKa = 7.63AGCAIGGDD9 pKa = 3.76DD10 pKa = 5.12SLNLSITRR18 pKa = 11.84PWLWADD24 pKa = 3.82DD25 pKa = 3.78KK26 pKa = 10.9VTSKK30 pKa = 10.32MYY32 pKa = 11.07GDD34 pKa = 4.25AGGNMSDD41 pKa = 4.1HH42 pKa = 6.64SVGGNDD48 pKa = 3.3VLSVYY53 pKa = 10.93LEE55 pKa = 4.15DD56 pKa = 3.89FQYY59 pKa = 11.3LRR61 pKa = 11.84VDD63 pKa = 3.41MFGDD67 pKa = 3.51AGGSLAGYY75 pKa = 10.5ASGGNDD81 pKa = 2.99TFSSDD86 pKa = 3.56THH88 pKa = 6.04WGHH91 pKa = 6.19IAVLIMYY98 pKa = 9.77GDD100 pKa = 3.95AGLNISGHH108 pKa = 5.94AFGGNDD114 pKa = 3.54VFNLGANVEE123 pKa = 4.41GNITCYY129 pKa = 10.87GDD131 pKa = 3.53AGGTMSGYY139 pKa = 9.15ATGGDD144 pKa = 3.85DD145 pKa = 4.27EE146 pKa = 4.75FTTAHH151 pKa = 5.05VRR153 pKa = 11.84EE154 pKa = 4.62TNFSIYY160 pKa = 10.64GDD162 pKa = 3.64AGGDD166 pKa = 3.32MSNYY170 pKa = 8.51ATGGNDD176 pKa = 2.85VFSANGEE183 pKa = 3.88NGYY186 pKa = 9.88VLFYY190 pKa = 11.3GDD192 pKa = 4.41AGGNMTGHH200 pKa = 6.53AQGGDD205 pKa = 3.33DD206 pKa = 4.48TYY208 pKa = 11.86NAGNDD213 pKa = 3.38ATTTFYY219 pKa = 11.65GDD221 pKa = 3.1AGGNMGDD228 pKa = 3.55RR229 pKa = 11.84TVGGNDD235 pKa = 3.31TFIGNAANFCGGVLAYY251 pKa = 10.55GDD253 pKa = 4.61ALTMAGSARR262 pKa = 11.84GGNDD266 pKa = 3.16TLVGASFSPTGNTLYY281 pKa = 11.21GDD283 pKa = 4.51ALTMSGNARR292 pKa = 11.84GGNDD296 pKa = 3.39TLIGGDD302 pKa = 3.63GVAAISGKK310 pKa = 9.88YY311 pKa = 9.37EE312 pKa = 4.38SILIGDD318 pKa = 4.84AKK320 pKa = 9.93TLSGNARR327 pKa = 11.84GGDD330 pKa = 3.54DD331 pKa = 3.62TLISGGGSDD340 pKa = 5.86DD341 pKa = 3.05MWGDD345 pKa = 3.18ARR347 pKa = 11.84VIGDD351 pKa = 3.75SARR354 pKa = 11.84GGDD357 pKa = 3.47DD358 pKa = 3.16RR359 pKa = 11.84FVFKK363 pKa = 10.86AGNGHH368 pKa = 7.08DD369 pKa = 4.37AIEE372 pKa = 5.24DD373 pKa = 4.01FGQGQAGSKK382 pKa = 9.24WGTDD386 pKa = 3.7HH387 pKa = 7.49IDD389 pKa = 3.17VSALGIQSFGALNISAFDD407 pKa = 3.89TATHH411 pKa = 6.71TSTLTFSAGNDD422 pKa = 3.47VVVHH426 pKa = 6.12SMLALRR432 pKa = 11.84AQDD435 pKa = 4.41FIFGG439 pKa = 3.78
MM1 pKa = 7.63AGCAIGGDD9 pKa = 3.76DD10 pKa = 5.12SLNLSITRR18 pKa = 11.84PWLWADD24 pKa = 3.82DD25 pKa = 3.78KK26 pKa = 10.9VTSKK30 pKa = 10.32MYY32 pKa = 11.07GDD34 pKa = 4.25AGGNMSDD41 pKa = 4.1HH42 pKa = 6.64SVGGNDD48 pKa = 3.3VLSVYY53 pKa = 10.93LEE55 pKa = 4.15DD56 pKa = 3.89FQYY59 pKa = 11.3LRR61 pKa = 11.84VDD63 pKa = 3.41MFGDD67 pKa = 3.51AGGSLAGYY75 pKa = 10.5ASGGNDD81 pKa = 2.99TFSSDD86 pKa = 3.56THH88 pKa = 6.04WGHH91 pKa = 6.19IAVLIMYY98 pKa = 9.77GDD100 pKa = 3.95AGLNISGHH108 pKa = 5.94AFGGNDD114 pKa = 3.54VFNLGANVEE123 pKa = 4.41GNITCYY129 pKa = 10.87GDD131 pKa = 3.53AGGTMSGYY139 pKa = 9.15ATGGDD144 pKa = 3.85DD145 pKa = 4.27EE146 pKa = 4.75FTTAHH151 pKa = 5.05VRR153 pKa = 11.84EE154 pKa = 4.62TNFSIYY160 pKa = 10.64GDD162 pKa = 3.64AGGDD166 pKa = 3.32MSNYY170 pKa = 8.51ATGGNDD176 pKa = 2.85VFSANGEE183 pKa = 3.88NGYY186 pKa = 9.88VLFYY190 pKa = 11.3GDD192 pKa = 4.41AGGNMTGHH200 pKa = 6.53AQGGDD205 pKa = 3.33DD206 pKa = 4.48TYY208 pKa = 11.86NAGNDD213 pKa = 3.38ATTTFYY219 pKa = 11.65GDD221 pKa = 3.1AGGNMGDD228 pKa = 3.55RR229 pKa = 11.84TVGGNDD235 pKa = 3.31TFIGNAANFCGGVLAYY251 pKa = 10.55GDD253 pKa = 4.61ALTMAGSARR262 pKa = 11.84GGNDD266 pKa = 3.16TLVGASFSPTGNTLYY281 pKa = 11.21GDD283 pKa = 4.51ALTMSGNARR292 pKa = 11.84GGNDD296 pKa = 3.39TLIGGDD302 pKa = 3.63GVAAISGKK310 pKa = 9.88YY311 pKa = 9.37EE312 pKa = 4.38SILIGDD318 pKa = 4.84AKK320 pKa = 9.93TLSGNARR327 pKa = 11.84GGDD330 pKa = 3.54DD331 pKa = 3.62TLISGGGSDD340 pKa = 5.86DD341 pKa = 3.05MWGDD345 pKa = 3.18ARR347 pKa = 11.84VIGDD351 pKa = 3.75SARR354 pKa = 11.84GGDD357 pKa = 3.47DD358 pKa = 3.16RR359 pKa = 11.84FVFKK363 pKa = 10.86AGNGHH368 pKa = 7.08DD369 pKa = 4.37AIEE372 pKa = 5.24DD373 pKa = 4.01FGQGQAGSKK382 pKa = 9.24WGTDD386 pKa = 3.7HH387 pKa = 7.49IDD389 pKa = 3.17VSALGIQSFGALNISAFDD407 pKa = 3.89TATHH411 pKa = 6.71TSTLTFSAGNDD422 pKa = 3.47VVVHH426 pKa = 6.12SMLALRR432 pKa = 11.84AQDD435 pKa = 4.41FIFGG439 pKa = 3.78
Molecular weight: 44.55 kDa
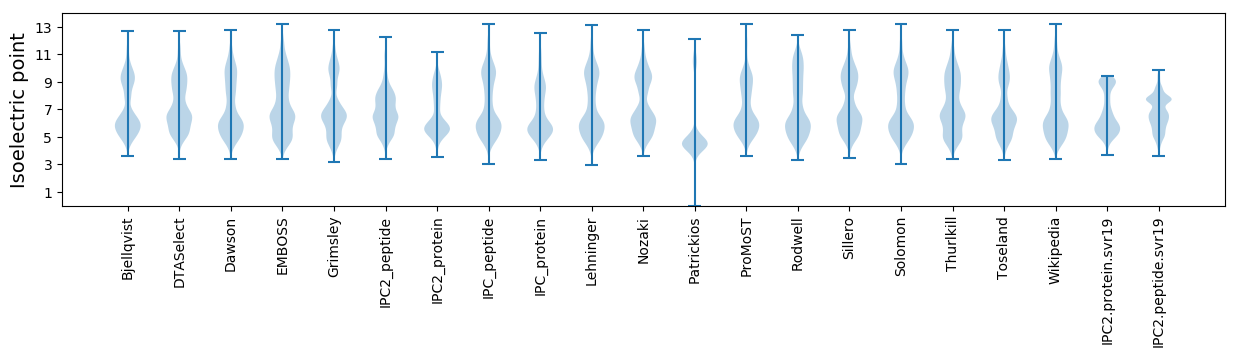
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S6N304|A0A2S6N304_9RHIZ Uncharacterized protein OS=Rhodoblastus sphagnicola OX=333368 GN=CCR94_16425 PE=4 SV=1
MM1 pKa = 7.93IPIPPKK7 pKa = 9.81NRR9 pKa = 11.84PAVRR13 pKa = 11.84SHH15 pKa = 4.52FTGRR19 pKa = 11.84GGFRR23 pKa = 11.84RR24 pKa = 11.84VLVQKK29 pKa = 10.56ARR31 pKa = 11.84RR32 pKa = 11.84SPRR35 pKa = 11.84LRR37 pKa = 11.84SNIARR42 pKa = 11.84RR43 pKa = 11.84FSAQAPVQHH52 pKa = 6.56GAAIICRR59 pKa = 11.84AKK61 pKa = 10.5SRR63 pKa = 11.84PQGAA67 pKa = 3.36
MM1 pKa = 7.93IPIPPKK7 pKa = 9.81NRR9 pKa = 11.84PAVRR13 pKa = 11.84SHH15 pKa = 4.52FTGRR19 pKa = 11.84GGFRR23 pKa = 11.84RR24 pKa = 11.84VLVQKK29 pKa = 10.56ARR31 pKa = 11.84RR32 pKa = 11.84SPRR35 pKa = 11.84LRR37 pKa = 11.84SNIARR42 pKa = 11.84RR43 pKa = 11.84FSAQAPVQHH52 pKa = 6.56GAAIICRR59 pKa = 11.84AKK61 pKa = 10.5SRR63 pKa = 11.84PQGAA67 pKa = 3.36
Molecular weight: 7.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1435672 |
25 |
4372 |
311.0 |
33.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.729 ± 0.058 | 0.961 ± 0.014 |
5.67 ± 0.027 | 5.613 ± 0.044 |
3.9 ± 0.027 | 8.383 ± 0.053 |
1.941 ± 0.018 | 4.885 ± 0.024 |
3.516 ± 0.03 | 10.311 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.124 ± 0.017 | 2.653 ± 0.025 |
5.188 ± 0.034 | 3.087 ± 0.021 |
7.264 ± 0.046 | 5.497 ± 0.038 |
4.868 ± 0.036 | 7.076 ± 0.027 |
1.24 ± 0.015 | 2.093 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
