
Coregonus sp. balchen
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha; Protacanthopterygii; Salmoniformes; Salmonidae; Coregoninae;
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
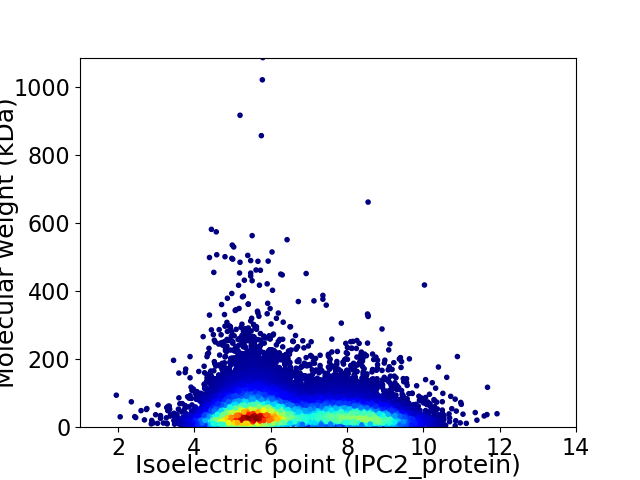
Virtual 2D-PAGE plot for 41641 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6F9CRJ2|A0A6F9CRJ2_9TELE RING-type domain-containing protein OS=Coregonus sp. 'balchen' OX=861768 GN=CSTEINMANNI_LOCUS4562867 PE=4 SV=1
MM1 pKa = 7.97EE2 pKa = 5.72DD3 pKa = 3.01NCRR6 pKa = 11.84LVPNKK11 pKa = 10.09DD12 pKa = 3.47QQNSDD17 pKa = 3.02TDD19 pKa = 4.05SYY21 pKa = 12.13GDD23 pKa = 4.06ACDD26 pKa = 3.63NCPNVPNSSQKK37 pKa = 9.86DD38 pKa = 3.41TDD40 pKa = 3.83GNGAGDD46 pKa = 3.66TCDD49 pKa = 3.98NDD51 pKa = 3.16IDD53 pKa = 4.83GDD55 pKa = 4.36GIPNVLDD62 pKa = 3.65NCPKK66 pKa = 10.25VPNPMQTDD74 pKa = 3.36RR75 pKa = 11.84DD76 pKa = 3.6GDD78 pKa = 4.04RR79 pKa = 11.84VGDD82 pKa = 4.0ACDD85 pKa = 3.35SCPEE89 pKa = 4.23VNDD92 pKa = 4.05PMQSDD97 pKa = 3.79MDD99 pKa = 4.0NDD101 pKa = 4.09LVGDD105 pKa = 3.95VCDD108 pKa = 4.47TNQDD112 pKa = 3.17MDD114 pKa = 4.21GDD116 pKa = 4.14GHH118 pKa = 6.78QDD120 pKa = 3.62SRR122 pKa = 11.84DD123 pKa = 3.61NCPDD127 pKa = 3.27VPNSSQLDD135 pKa = 3.58SDD137 pKa = 3.72NDD139 pKa = 4.86GIGDD143 pKa = 3.95EE144 pKa = 5.45CDD146 pKa = 5.19DD147 pKa = 5.83DD148 pKa = 6.4DD149 pKa = 7.03DD150 pKa = 6.24NDD152 pKa = 5.35GIPDD156 pKa = 3.89TRR158 pKa = 11.84PPGPDD163 pKa = 2.91NCRR166 pKa = 11.84LIPNPSQTDD175 pKa = 3.2TDD177 pKa = 4.28GNGVGDD183 pKa = 3.63MCEE186 pKa = 3.88NDD188 pKa = 3.53FDD190 pKa = 4.34NDD192 pKa = 4.02SVIDD196 pKa = 4.89LIDD199 pKa = 3.48VCPEE203 pKa = 3.75SAEE206 pKa = 3.87VTMTDD211 pKa = 3.44FRR213 pKa = 11.84AYY215 pKa = 8.36QTVILDD221 pKa = 3.91PEE223 pKa = 5.2GDD225 pKa = 3.69AQIDD229 pKa = 4.02PNWIVLNRR237 pKa = 11.84GMEE240 pKa = 4.2IVQTMNSDD248 pKa = 3.24PGLAIGYY255 pKa = 6.53TAFNGVDD262 pKa = 3.99FEE264 pKa = 4.94GTFHH268 pKa = 7.41INTATDD274 pKa = 3.36DD275 pKa = 4.34DD276 pKa = 4.73YY277 pKa = 12.04VGFIFGYY284 pKa = 9.23QDD286 pKa = 2.88SSSFYY291 pKa = 10.16VYY293 pKa = 10.6LRR295 pKa = 11.84NALWHH300 pKa = 6.31TGDD303 pKa = 3.47TPGEE307 pKa = 4.12VTLLWKK313 pKa = 10.49DD314 pKa = 3.4PRR316 pKa = 11.84NVGWKK321 pKa = 10.34DD322 pKa = 2.99KK323 pKa = 10.84TSYY326 pKa = 10.02RR327 pKa = 11.84WHH329 pKa = 6.91LSHH332 pKa = 7.13RR333 pKa = 11.84PQVGYY338 pKa = 10.62INVRR342 pKa = 11.84LYY344 pKa = 11.3EE345 pKa = 4.1GMTLVADD352 pKa = 3.69SGMVVDD358 pKa = 3.88TSMRR362 pKa = 11.84GGRR365 pKa = 11.84LGVFCFSQEE374 pKa = 3.83QIIWSNLGYY383 pKa = 10.17RR384 pKa = 11.84CNDD387 pKa = 3.6TVPEE391 pKa = 4.4DD392 pKa = 3.44YY393 pKa = 10.91EE394 pKa = 4.55FYY396 pKa = 10.6RR397 pKa = 11.84KK398 pKa = 9.56QIHH401 pKa = 5.42VRR403 pKa = 11.84VV404 pKa = 3.77
MM1 pKa = 7.97EE2 pKa = 5.72DD3 pKa = 3.01NCRR6 pKa = 11.84LVPNKK11 pKa = 10.09DD12 pKa = 3.47QQNSDD17 pKa = 3.02TDD19 pKa = 4.05SYY21 pKa = 12.13GDD23 pKa = 4.06ACDD26 pKa = 3.63NCPNVPNSSQKK37 pKa = 9.86DD38 pKa = 3.41TDD40 pKa = 3.83GNGAGDD46 pKa = 3.66TCDD49 pKa = 3.98NDD51 pKa = 3.16IDD53 pKa = 4.83GDD55 pKa = 4.36GIPNVLDD62 pKa = 3.65NCPKK66 pKa = 10.25VPNPMQTDD74 pKa = 3.36RR75 pKa = 11.84DD76 pKa = 3.6GDD78 pKa = 4.04RR79 pKa = 11.84VGDD82 pKa = 4.0ACDD85 pKa = 3.35SCPEE89 pKa = 4.23VNDD92 pKa = 4.05PMQSDD97 pKa = 3.79MDD99 pKa = 4.0NDD101 pKa = 4.09LVGDD105 pKa = 3.95VCDD108 pKa = 4.47TNQDD112 pKa = 3.17MDD114 pKa = 4.21GDD116 pKa = 4.14GHH118 pKa = 6.78QDD120 pKa = 3.62SRR122 pKa = 11.84DD123 pKa = 3.61NCPDD127 pKa = 3.27VPNSSQLDD135 pKa = 3.58SDD137 pKa = 3.72NDD139 pKa = 4.86GIGDD143 pKa = 3.95EE144 pKa = 5.45CDD146 pKa = 5.19DD147 pKa = 5.83DD148 pKa = 6.4DD149 pKa = 7.03DD150 pKa = 6.24NDD152 pKa = 5.35GIPDD156 pKa = 3.89TRR158 pKa = 11.84PPGPDD163 pKa = 2.91NCRR166 pKa = 11.84LIPNPSQTDD175 pKa = 3.2TDD177 pKa = 4.28GNGVGDD183 pKa = 3.63MCEE186 pKa = 3.88NDD188 pKa = 3.53FDD190 pKa = 4.34NDD192 pKa = 4.02SVIDD196 pKa = 4.89LIDD199 pKa = 3.48VCPEE203 pKa = 3.75SAEE206 pKa = 3.87VTMTDD211 pKa = 3.44FRR213 pKa = 11.84AYY215 pKa = 8.36QTVILDD221 pKa = 3.91PEE223 pKa = 5.2GDD225 pKa = 3.69AQIDD229 pKa = 4.02PNWIVLNRR237 pKa = 11.84GMEE240 pKa = 4.2IVQTMNSDD248 pKa = 3.24PGLAIGYY255 pKa = 6.53TAFNGVDD262 pKa = 3.99FEE264 pKa = 4.94GTFHH268 pKa = 7.41INTATDD274 pKa = 3.36DD275 pKa = 4.34DD276 pKa = 4.73YY277 pKa = 12.04VGFIFGYY284 pKa = 9.23QDD286 pKa = 2.88SSSFYY291 pKa = 10.16VYY293 pKa = 10.6LRR295 pKa = 11.84NALWHH300 pKa = 6.31TGDD303 pKa = 3.47TPGEE307 pKa = 4.12VTLLWKK313 pKa = 10.49DD314 pKa = 3.4PRR316 pKa = 11.84NVGWKK321 pKa = 10.34DD322 pKa = 2.99KK323 pKa = 10.84TSYY326 pKa = 10.02RR327 pKa = 11.84WHH329 pKa = 6.91LSHH332 pKa = 7.13RR333 pKa = 11.84PQVGYY338 pKa = 10.62INVRR342 pKa = 11.84LYY344 pKa = 11.3EE345 pKa = 4.1GMTLVADD352 pKa = 3.69SGMVVDD358 pKa = 3.88TSMRR362 pKa = 11.84GGRR365 pKa = 11.84LGVFCFSQEE374 pKa = 3.83QIIWSNLGYY383 pKa = 10.17RR384 pKa = 11.84CNDD387 pKa = 3.6TVPEE391 pKa = 4.4DD392 pKa = 3.44YY393 pKa = 10.91EE394 pKa = 4.55FYY396 pKa = 10.6RR397 pKa = 11.84KK398 pKa = 9.56QIHH401 pKa = 5.42VRR403 pKa = 11.84VV404 pKa = 3.77
Molecular weight: 44.75 kDa
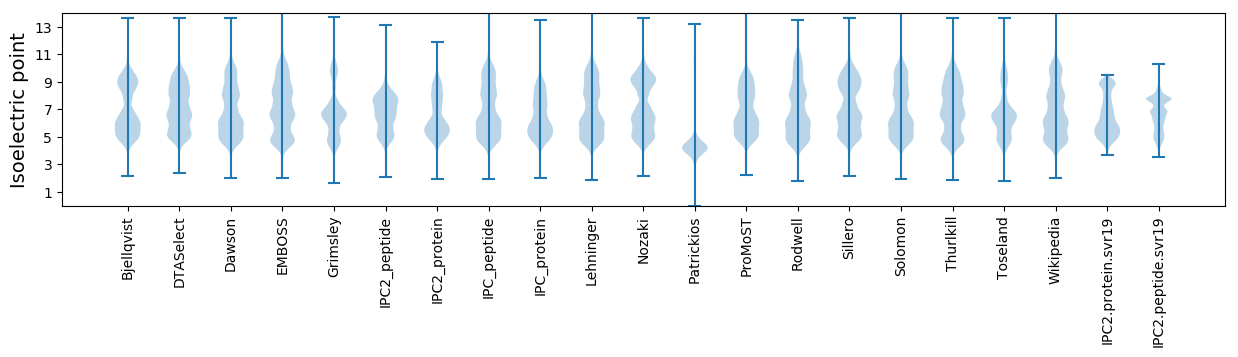
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6F8ZXA9|A0A6F8ZXA9_9TELE Uncharacterized protein (Fragment) OS=Coregonus sp. 'balchen' OX=861768 GN=CSTEINMANNI_LOCUS508758 PE=4 SV=1
TT1 pKa = 7.13LRR3 pKa = 11.84NPGLIINSPVTLRR16 pKa = 11.84NPGLIINSPVTLRR29 pKa = 11.84NPGLIINSPVTLRR42 pKa = 11.84NPGLIINSPVTLRR55 pKa = 11.84NPGLIINPPVTLRR68 pKa = 11.84NPGLIINSPVTLRR81 pKa = 11.84NPGLITNSPVTLRR94 pKa = 11.84NPGLIINPPVTIRR107 pKa = 11.84NPGLITNPPKK117 pKa = 10.27PWTNHH122 pKa = 4.38QPSITLRR129 pKa = 11.84NPGLITNPPVTLRR142 pKa = 11.84NPGLIINPPVTLRR155 pKa = 11.84NPGLIINSPVTLRR168 pKa = 11.84NPGLIINPPVTLRR181 pKa = 11.84NPGLIINPPLPSEE194 pKa = 4.15NPGLIINPPVTLRR207 pKa = 11.84NPGLIINPPVTLRR220 pKa = 11.84NPGLIINSPVTLRR233 pKa = 11.84NPGLIINPPVTLRR246 pKa = 11.84NPGLIINPPVTLRR259 pKa = 11.84NPGLITNPPVTLRR272 pKa = 11.84NPGLIINPPVTLRR285 pKa = 11.84NPGLIINPPVTLRR298 pKa = 11.84NPGLITNPPVTLRR311 pKa = 11.84NPGLIINPPVTLRR324 pKa = 11.84NPGLIINPPVTLRR337 pKa = 11.84NPGLIINPPVTLRR350 pKa = 11.84NPGLIINPPVTLRR363 pKa = 11.84NPGLIINPP371 pKa = 3.92
TT1 pKa = 7.13LRR3 pKa = 11.84NPGLIINSPVTLRR16 pKa = 11.84NPGLIINSPVTLRR29 pKa = 11.84NPGLIINSPVTLRR42 pKa = 11.84NPGLIINSPVTLRR55 pKa = 11.84NPGLIINPPVTLRR68 pKa = 11.84NPGLIINSPVTLRR81 pKa = 11.84NPGLITNSPVTLRR94 pKa = 11.84NPGLIINPPVTIRR107 pKa = 11.84NPGLITNPPKK117 pKa = 10.27PWTNHH122 pKa = 4.38QPSITLRR129 pKa = 11.84NPGLITNPPVTLRR142 pKa = 11.84NPGLIINPPVTLRR155 pKa = 11.84NPGLIINSPVTLRR168 pKa = 11.84NPGLIINPPVTLRR181 pKa = 11.84NPGLIINPPLPSEE194 pKa = 4.15NPGLIINPPVTLRR207 pKa = 11.84NPGLIINPPVTLRR220 pKa = 11.84NPGLIINSPVTLRR233 pKa = 11.84NPGLIINPPVTLRR246 pKa = 11.84NPGLIINPPVTLRR259 pKa = 11.84NPGLITNPPVTLRR272 pKa = 11.84NPGLIINPPVTLRR285 pKa = 11.84NPGLIINPPVTLRR298 pKa = 11.84NPGLITNPPVTLRR311 pKa = 11.84NPGLIINPPVTLRR324 pKa = 11.84NPGLIINPPVTLRR337 pKa = 11.84NPGLIINPPVTLRR350 pKa = 11.84NPGLIINPPVTLRR363 pKa = 11.84NPGLIINPP371 pKa = 3.92
Molecular weight: 39.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
18652202 |
9 |
12605 |
447.9 |
49.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.329 ± 0.013 | 2.203 ± 0.011 |
5.084 ± 0.01 | 6.927 ± 0.019 |
3.293 ± 0.008 | 6.763 ± 0.016 |
2.743 ± 0.009 | 4.111 ± 0.01 |
5.275 ± 0.016 | 9.404 ± 0.019 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.46 ± 0.005 | 3.705 ± 0.008 |
6.234 ± 0.02 | 4.78 ± 0.013 |
5.797 ± 0.012 | 8.705 ± 0.017 |
5.967 ± 0.016 | 6.352 ± 0.013 |
1.165 ± 0.005 | 2.703 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
