
Capybara microvirus Cap1_SP_116
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
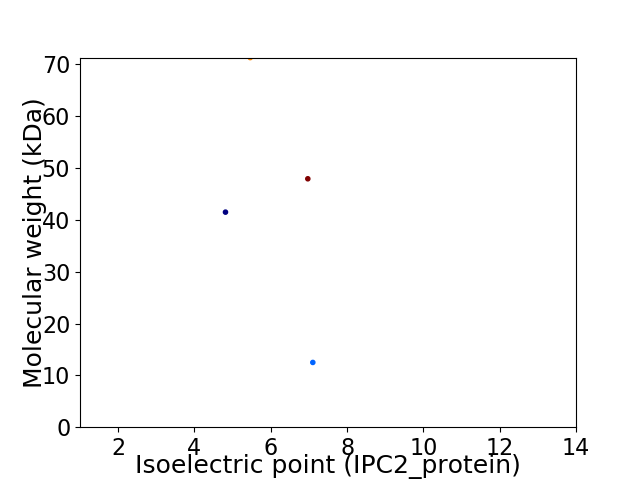
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
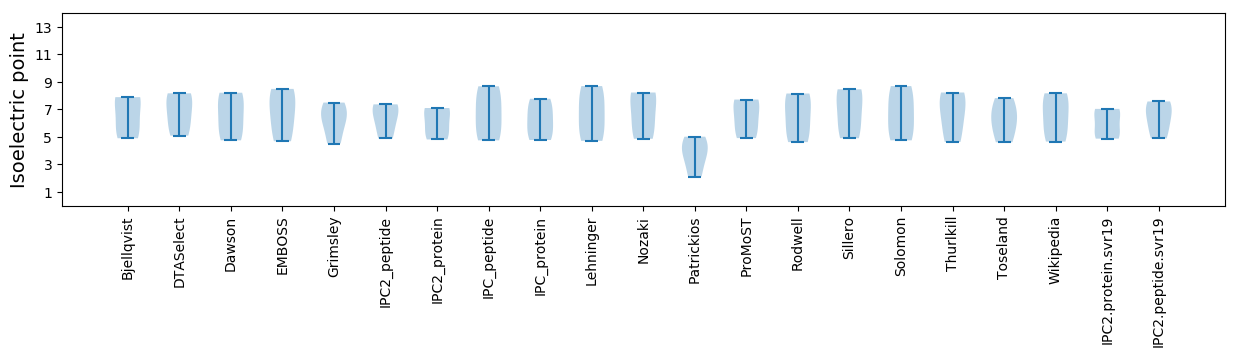
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVP0|A0A4V1FVP0_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_116 OX=2584809 PE=4 SV=1
MM1 pKa = 8.11PDD3 pKa = 3.69LSNHH7 pKa = 6.7AGSQVTPNPVTSGPYY22 pKa = 9.76SVVNTADD29 pKa = 3.86QLSGWNRR36 pKa = 11.84LTDD39 pKa = 3.42WLGVTNHH46 pKa = 5.99QGQINNQLALQQSLLDD62 pKa = 4.7DD63 pKa = 3.53EE64 pKa = 5.46RR65 pKa = 11.84AYY67 pKa = 11.25NDD69 pKa = 3.35PAAQVEE75 pKa = 4.27RR76 pKa = 11.84LRR78 pKa = 11.84NAGINPDD85 pKa = 3.6LLGGVSSYY93 pKa = 11.44EE94 pKa = 4.11STGQGANPLSQPTGDD109 pKa = 3.58FGNFILGVAQTFSGVASGILDD130 pKa = 4.08LQSKK134 pKa = 10.46ALDD137 pKa = 3.64NDD139 pKa = 3.46IKK141 pKa = 10.74RR142 pKa = 11.84ASLFDD147 pKa = 3.51TEE149 pKa = 5.41SILNRR154 pKa = 11.84IASDD158 pKa = 3.48RR159 pKa = 11.84SISADD164 pKa = 3.79FIEE167 pKa = 5.59AMKK170 pKa = 10.73DD171 pKa = 3.34PNRR174 pKa = 11.84LKK176 pKa = 10.66PAVSLDD182 pKa = 3.28QYY184 pKa = 11.62VNLSTIRR191 pKa = 11.84SKK193 pKa = 10.8KK194 pKa = 8.03VRR196 pKa = 11.84KK197 pKa = 9.28AVEE200 pKa = 3.87DD201 pKa = 3.48NFYY204 pKa = 11.35SKK206 pKa = 10.77LYY208 pKa = 10.61SDD210 pKa = 6.04DD211 pKa = 4.12FIQRR215 pKa = 11.84INGLLTGVNKK225 pKa = 10.01SEE227 pKa = 4.33RR228 pKa = 11.84DD229 pKa = 3.45FSEE232 pKa = 4.31SVAGLRR238 pKa = 11.84NPSKK242 pKa = 9.73TADD245 pKa = 3.64SLIDD249 pKa = 3.52ALAEE253 pKa = 4.19FMSDD257 pKa = 3.3AYY259 pKa = 7.52TTQMSANKK267 pKa = 10.14AEE269 pKa = 4.13RR270 pKa = 11.84HH271 pKa = 6.28HH272 pKa = 6.77NDD274 pKa = 2.94ASISQDD280 pKa = 3.63DD281 pKa = 4.16YY282 pKa = 11.99NKK284 pKa = 9.92MVTDD288 pKa = 4.88ALDD291 pKa = 3.78PSNEE295 pKa = 3.9SDD297 pKa = 3.94AITAGFEE304 pKa = 3.89ASEE307 pKa = 4.26TSKK310 pKa = 10.86KK311 pKa = 9.68KK312 pKa = 10.42QSNIDD317 pKa = 3.55SVIEE321 pKa = 3.98EE322 pKa = 4.38FTGKK326 pKa = 8.8MKK328 pKa = 10.83KK329 pKa = 9.82KK330 pKa = 9.27DD331 pKa = 3.65TWWSDD336 pKa = 3.64LILAIMAMYY345 pKa = 10.32QSGAFDD351 pKa = 3.22SFGFSRR357 pKa = 11.84SSQSSFTPRR366 pKa = 11.84GGSQEE371 pKa = 3.49RR372 pKa = 11.84SSFNFHH378 pKa = 6.92LL379 pKa = 4.6
MM1 pKa = 8.11PDD3 pKa = 3.69LSNHH7 pKa = 6.7AGSQVTPNPVTSGPYY22 pKa = 9.76SVVNTADD29 pKa = 3.86QLSGWNRR36 pKa = 11.84LTDD39 pKa = 3.42WLGVTNHH46 pKa = 5.99QGQINNQLALQQSLLDD62 pKa = 4.7DD63 pKa = 3.53EE64 pKa = 5.46RR65 pKa = 11.84AYY67 pKa = 11.25NDD69 pKa = 3.35PAAQVEE75 pKa = 4.27RR76 pKa = 11.84LRR78 pKa = 11.84NAGINPDD85 pKa = 3.6LLGGVSSYY93 pKa = 11.44EE94 pKa = 4.11STGQGANPLSQPTGDD109 pKa = 3.58FGNFILGVAQTFSGVASGILDD130 pKa = 4.08LQSKK134 pKa = 10.46ALDD137 pKa = 3.64NDD139 pKa = 3.46IKK141 pKa = 10.74RR142 pKa = 11.84ASLFDD147 pKa = 3.51TEE149 pKa = 5.41SILNRR154 pKa = 11.84IASDD158 pKa = 3.48RR159 pKa = 11.84SISADD164 pKa = 3.79FIEE167 pKa = 5.59AMKK170 pKa = 10.73DD171 pKa = 3.34PNRR174 pKa = 11.84LKK176 pKa = 10.66PAVSLDD182 pKa = 3.28QYY184 pKa = 11.62VNLSTIRR191 pKa = 11.84SKK193 pKa = 10.8KK194 pKa = 8.03VRR196 pKa = 11.84KK197 pKa = 9.28AVEE200 pKa = 3.87DD201 pKa = 3.48NFYY204 pKa = 11.35SKK206 pKa = 10.77LYY208 pKa = 10.61SDD210 pKa = 6.04DD211 pKa = 4.12FIQRR215 pKa = 11.84INGLLTGVNKK225 pKa = 10.01SEE227 pKa = 4.33RR228 pKa = 11.84DD229 pKa = 3.45FSEE232 pKa = 4.31SVAGLRR238 pKa = 11.84NPSKK242 pKa = 9.73TADD245 pKa = 3.64SLIDD249 pKa = 3.52ALAEE253 pKa = 4.19FMSDD257 pKa = 3.3AYY259 pKa = 7.52TTQMSANKK267 pKa = 10.14AEE269 pKa = 4.13RR270 pKa = 11.84HH271 pKa = 6.28HH272 pKa = 6.77NDD274 pKa = 2.94ASISQDD280 pKa = 3.63DD281 pKa = 4.16YY282 pKa = 11.99NKK284 pKa = 9.92MVTDD288 pKa = 4.88ALDD291 pKa = 3.78PSNEE295 pKa = 3.9SDD297 pKa = 3.94AITAGFEE304 pKa = 3.89ASEE307 pKa = 4.26TSKK310 pKa = 10.86KK311 pKa = 9.68KK312 pKa = 10.42QSNIDD317 pKa = 3.55SVIEE321 pKa = 3.98EE322 pKa = 4.38FTGKK326 pKa = 8.8MKK328 pKa = 10.83KK329 pKa = 9.82KK330 pKa = 9.27DD331 pKa = 3.65TWWSDD336 pKa = 3.64LILAIMAMYY345 pKa = 10.32QSGAFDD351 pKa = 3.22SFGFSRR357 pKa = 11.84SSQSSFTPRR366 pKa = 11.84GGSQEE371 pKa = 3.49RR372 pKa = 11.84SSFNFHH378 pKa = 6.92LL379 pKa = 4.6
Molecular weight: 41.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W461|A0A4P8W461_9VIRU Replication initiator protein OS=Capybara microvirus Cap1_SP_116 OX=2584809 PE=4 SV=1
MM1 pKa = 7.64SILALILSLVSLIRR15 pKa = 11.84TGRR18 pKa = 11.84NHH20 pKa = 7.12RR21 pKa = 11.84DD22 pKa = 2.93VSRR25 pKa = 11.84LVDD28 pKa = 3.77TVAHH32 pKa = 6.18IEE34 pKa = 3.8EE35 pKa = 4.75SIRR38 pKa = 11.84VNDD41 pKa = 3.5QAFVDD46 pKa = 3.81ACKK49 pKa = 10.75SFTEE53 pKa = 4.6SIEE56 pKa = 4.06KK57 pKa = 10.26LDD59 pKa = 3.73YY60 pKa = 11.07ALVMLEE66 pKa = 4.8RR67 pKa = 11.84YY68 pKa = 6.69TLPSGSSSLIVTGPRR83 pKa = 11.84SKK85 pKa = 10.01VTTTDD90 pKa = 3.06SEE92 pKa = 5.49SNMSVSDD99 pKa = 3.88SRR101 pKa = 11.84NQSSDD106 pKa = 3.04KK107 pKa = 10.62KK108 pKa = 10.99SKK110 pKa = 10.69NKK112 pKa = 9.97NN113 pKa = 3.06
MM1 pKa = 7.64SILALILSLVSLIRR15 pKa = 11.84TGRR18 pKa = 11.84NHH20 pKa = 7.12RR21 pKa = 11.84DD22 pKa = 2.93VSRR25 pKa = 11.84LVDD28 pKa = 3.77TVAHH32 pKa = 6.18IEE34 pKa = 3.8EE35 pKa = 4.75SIRR38 pKa = 11.84VNDD41 pKa = 3.5QAFVDD46 pKa = 3.81ACKK49 pKa = 10.75SFTEE53 pKa = 4.6SIEE56 pKa = 4.06KK57 pKa = 10.26LDD59 pKa = 3.73YY60 pKa = 11.07ALVMLEE66 pKa = 4.8RR67 pKa = 11.84YY68 pKa = 6.69TLPSGSSSLIVTGPRR83 pKa = 11.84SKK85 pKa = 10.01VTTTDD90 pKa = 3.06SEE92 pKa = 5.49SNMSVSDD99 pKa = 3.88SRR101 pKa = 11.84NQSSDD106 pKa = 3.04KK107 pKa = 10.62KK108 pKa = 10.99SKK110 pKa = 10.69NKK112 pKa = 9.97NN113 pKa = 3.06
Molecular weight: 12.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1529 |
113 |
630 |
382.3 |
43.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.971 ± 1.451 | 1.57 ± 0.66 |
7.456 ± 0.709 | 3.924 ± 0.327 |
4.84 ± 0.479 | 5.298 ± 0.449 |
2.42 ± 0.371 | 5.298 ± 0.529 |
4.971 ± 1.105 | 8.698 ± 0.199 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.962 ± 0.121 | 6.802 ± 0.563 |
4.186 ± 0.732 | 4.382 ± 0.568 |
4.971 ± 0.443 | 11.445 ± 1.123 |
4.774 ± 0.854 | 5.755 ± 0.533 |
1.046 ± 0.182 | 5.232 ± 1.088 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
