
Beihai narna-like virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.87
Get precalculated fractions of proteins
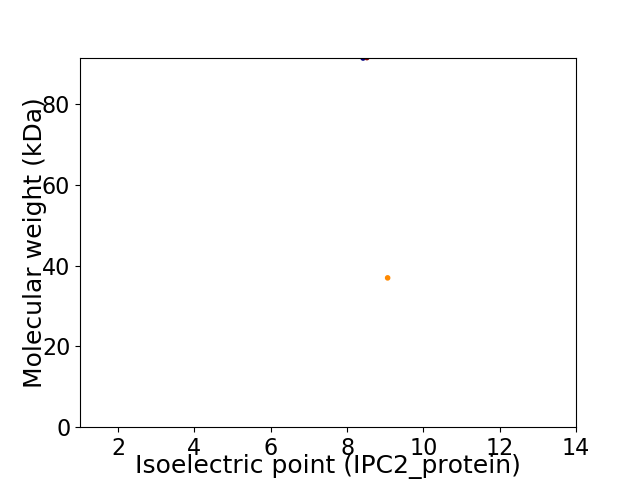
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KIK3|A0A1L3KIK3_9VIRU Uncharacterized protein OS=Beihai narna-like virus 9 OX=1922461 PE=4 SV=1
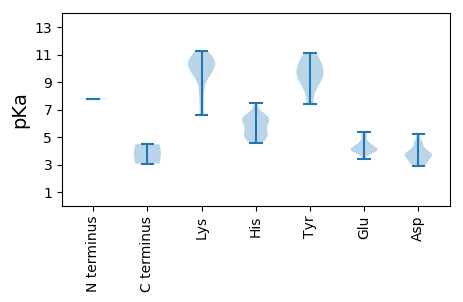
MM1 pKa = 7.79SPEE4 pKa = 4.66DD5 pKa = 4.59LPACLEE11 pKa = 3.92RR12 pKa = 11.84AARR15 pKa = 11.84ALAGRR20 pKa = 11.84QKK22 pKa = 9.39WLHH25 pKa = 5.1MKK27 pKa = 9.88HH28 pKa = 6.41RR29 pKa = 11.84SFVALTNGAYY39 pKa = 10.26FLRR42 pKa = 11.84SLVEE46 pKa = 3.96VLLDD50 pKa = 3.96ADD52 pKa = 4.9PIFLRR57 pKa = 11.84LKK59 pKa = 10.65LADD62 pKa = 3.7FWEE65 pKa = 4.7VFSILWPISPAEE77 pKa = 3.79QVAVLKK83 pKa = 10.81YY84 pKa = 8.47WVAWPMARR92 pKa = 11.84WLRR95 pKa = 11.84DD96 pKa = 3.01SDD98 pKa = 3.62ITSARR103 pKa = 11.84PPCLPPAPTSLDD115 pKa = 3.47FPLRR119 pKa = 11.84GPLRR123 pKa = 11.84KK124 pKa = 9.5HH125 pKa = 5.62FRR127 pKa = 11.84NMLASRR133 pKa = 11.84TSSVRR138 pKa = 11.84AGTLFTGILQGVKK151 pKa = 9.93RR152 pKa = 11.84GCAPVPEE159 pKa = 4.34EE160 pKa = 5.12FEE162 pKa = 4.27VLSCQKK168 pKa = 10.29HH169 pKa = 5.29KK170 pKa = 10.82KK171 pKa = 10.06ALSQPLEE178 pKa = 4.18FSHH181 pKa = 7.49GPEE184 pKa = 4.22FFAKK188 pKa = 10.44FQAIWGKK195 pKa = 8.6TNTEE199 pKa = 3.43NRR201 pKa = 11.84RR202 pKa = 11.84AALHH206 pKa = 5.76SRR208 pKa = 11.84WKK210 pKa = 10.57RR211 pKa = 11.84VFLDD215 pKa = 3.82RR216 pKa = 11.84KK217 pKa = 7.39WARR220 pKa = 11.84HH221 pKa = 4.59LHH223 pKa = 5.47SASNHH228 pKa = 5.18ASVEE232 pKa = 4.14RR233 pKa = 11.84MRR235 pKa = 11.84SEE237 pKa = 4.04GGRR240 pKa = 11.84VAEE243 pKa = 4.09VFHH246 pKa = 6.33VNRR249 pKa = 11.84ILIGEE254 pKa = 4.28DD255 pKa = 3.13RR256 pKa = 11.84DD257 pKa = 3.75PRR259 pKa = 11.84LVDD262 pKa = 3.99EE263 pKa = 5.4PPLLDD268 pKa = 3.14MFEE271 pKa = 4.66RR272 pKa = 11.84NGRR275 pKa = 11.84VVEE278 pKa = 4.0RR279 pKa = 11.84RR280 pKa = 11.84GWPLASYY287 pKa = 10.37RR288 pKa = 11.84QFLRR292 pKa = 11.84QAAGMCRR299 pKa = 11.84GRR301 pKa = 11.84DD302 pKa = 3.12LRR304 pKa = 11.84ARR306 pKa = 11.84VEE308 pKa = 4.11LCLEE312 pKa = 4.03PLKK315 pKa = 11.02CRR317 pKa = 11.84VITKK321 pKa = 10.26GEE323 pKa = 4.07SVPYY327 pKa = 9.8FVAQTFQKK335 pKa = 10.96SMWKK339 pKa = 10.17ALQEE343 pKa = 3.93TGAFKK348 pKa = 9.95LTSCPVDD355 pKa = 3.13ASMLYY360 pKa = 10.64GIEE363 pKa = 5.22LSTKK367 pKa = 10.68DD368 pKa = 3.78LDD370 pKa = 4.34LPFDD374 pKa = 3.36QWVSGDD380 pKa = 3.54YY381 pKa = 10.74SAATDD386 pKa = 3.88GLTLEE391 pKa = 5.1VNQCCLRR398 pKa = 11.84AMLDD402 pKa = 3.48AFQATPEE409 pKa = 3.93EE410 pKa = 4.57RR411 pKa = 11.84EE412 pKa = 4.16VCSKK416 pKa = 11.09VLGCHH421 pKa = 4.83EE422 pKa = 3.96VSYY425 pKa = 10.69PDD427 pKa = 4.53RR428 pKa = 11.84LDD430 pKa = 3.78PEE432 pKa = 4.96ANGLEE437 pKa = 4.37PFHH440 pKa = 6.33MLNGQLMGSVLSFPVLCAVNLAAYY464 pKa = 7.54WCALEE469 pKa = 5.16EE470 pKa = 4.0YY471 pKa = 9.57TGRR474 pKa = 11.84KK475 pKa = 8.16FKK477 pKa = 10.96KK478 pKa = 8.37EE479 pKa = 3.8QLPVLVNGDD488 pKa = 4.35DD489 pKa = 3.95ILFKK493 pKa = 11.24ANAAFYY499 pKa = 9.0EE500 pKa = 4.68VWKK503 pKa = 10.24KK504 pKa = 6.7WVKK507 pKa = 10.22RR508 pKa = 11.84AGFSLSLGKK517 pKa = 10.4NYY519 pKa = 9.9ISPNFITVNSEE530 pKa = 3.46SWLHH534 pKa = 6.17KK535 pKa = 9.5GGSYY539 pKa = 9.82FRR541 pKa = 11.84KK542 pKa = 10.14LPFLNCGLLLQEE554 pKa = 4.42AAGPYY559 pKa = 9.19KK560 pKa = 10.78VPLRR564 pKa = 11.84AEE566 pKa = 3.87TAEE569 pKa = 4.15RR570 pKa = 11.84PLIPKK575 pKa = 7.98LQWIIDD581 pKa = 3.77NCNNPARR588 pKa = 11.84AFDD591 pKa = 4.64RR592 pKa = 11.84IKK594 pKa = 10.27HH595 pKa = 4.76HH596 pKa = 5.75WRR598 pKa = 11.84RR599 pKa = 11.84SINVHH604 pKa = 4.87TEE606 pKa = 4.04FGRR609 pKa = 11.84FNLCAPMEE617 pKa = 4.37LGGCGLKK624 pKa = 10.61LPEE627 pKa = 4.23AVRR630 pKa = 11.84PAVNFTAFQCLLAGRR645 pKa = 11.84SHH647 pKa = 6.45QSYY650 pKa = 11.11KK651 pKa = 10.72NLEE654 pKa = 4.05GTDD657 pKa = 2.95IRR659 pKa = 11.84DD660 pKa = 3.5RR661 pKa = 11.84PKK663 pKa = 10.49TGFEE667 pKa = 4.26RR668 pKa = 11.84ISVAEE673 pKa = 4.01VSKK676 pKa = 10.93SINPLPTEE684 pKa = 4.15SRR686 pKa = 11.84LGTAVLRR693 pKa = 11.84SPFEE697 pKa = 3.81PVRR700 pKa = 11.84GEE702 pKa = 3.94LEE704 pKa = 4.0VRR706 pKa = 11.84FEE708 pKa = 5.31DD709 pKa = 5.22PIACLRR715 pKa = 11.84VAGEE719 pKa = 4.38LNTAQTASDD728 pKa = 3.59QEE730 pKa = 4.23RR731 pKa = 11.84PVYY734 pKa = 9.61VLKK737 pKa = 10.74RR738 pKa = 11.84LSRR741 pKa = 11.84KK742 pKa = 9.47RR743 pKa = 11.84LGAVFSTGAKK753 pKa = 6.67ITKK756 pKa = 9.34PFLFPFEE763 pKa = 4.07VRR765 pKa = 11.84KK766 pKa = 9.6QLTRR770 pKa = 11.84GPTDD774 pKa = 3.77DD775 pKa = 4.7ASPPAMPEE783 pKa = 4.54DD784 pKa = 3.62IAEE787 pKa = 4.3MATEE791 pKa = 4.6LCKK794 pKa = 10.52SAPKK798 pKa = 9.65PSCGGGTSRR807 pKa = 11.84KK808 pKa = 6.6WW809 pKa = 3.06
MM1 pKa = 7.79SPEE4 pKa = 4.66DD5 pKa = 4.59LPACLEE11 pKa = 3.92RR12 pKa = 11.84AARR15 pKa = 11.84ALAGRR20 pKa = 11.84QKK22 pKa = 9.39WLHH25 pKa = 5.1MKK27 pKa = 9.88HH28 pKa = 6.41RR29 pKa = 11.84SFVALTNGAYY39 pKa = 10.26FLRR42 pKa = 11.84SLVEE46 pKa = 3.96VLLDD50 pKa = 3.96ADD52 pKa = 4.9PIFLRR57 pKa = 11.84LKK59 pKa = 10.65LADD62 pKa = 3.7FWEE65 pKa = 4.7VFSILWPISPAEE77 pKa = 3.79QVAVLKK83 pKa = 10.81YY84 pKa = 8.47WVAWPMARR92 pKa = 11.84WLRR95 pKa = 11.84DD96 pKa = 3.01SDD98 pKa = 3.62ITSARR103 pKa = 11.84PPCLPPAPTSLDD115 pKa = 3.47FPLRR119 pKa = 11.84GPLRR123 pKa = 11.84KK124 pKa = 9.5HH125 pKa = 5.62FRR127 pKa = 11.84NMLASRR133 pKa = 11.84TSSVRR138 pKa = 11.84AGTLFTGILQGVKK151 pKa = 9.93RR152 pKa = 11.84GCAPVPEE159 pKa = 4.34EE160 pKa = 5.12FEE162 pKa = 4.27VLSCQKK168 pKa = 10.29HH169 pKa = 5.29KK170 pKa = 10.82KK171 pKa = 10.06ALSQPLEE178 pKa = 4.18FSHH181 pKa = 7.49GPEE184 pKa = 4.22FFAKK188 pKa = 10.44FQAIWGKK195 pKa = 8.6TNTEE199 pKa = 3.43NRR201 pKa = 11.84RR202 pKa = 11.84AALHH206 pKa = 5.76SRR208 pKa = 11.84WKK210 pKa = 10.57RR211 pKa = 11.84VFLDD215 pKa = 3.82RR216 pKa = 11.84KK217 pKa = 7.39WARR220 pKa = 11.84HH221 pKa = 4.59LHH223 pKa = 5.47SASNHH228 pKa = 5.18ASVEE232 pKa = 4.14RR233 pKa = 11.84MRR235 pKa = 11.84SEE237 pKa = 4.04GGRR240 pKa = 11.84VAEE243 pKa = 4.09VFHH246 pKa = 6.33VNRR249 pKa = 11.84ILIGEE254 pKa = 4.28DD255 pKa = 3.13RR256 pKa = 11.84DD257 pKa = 3.75PRR259 pKa = 11.84LVDD262 pKa = 3.99EE263 pKa = 5.4PPLLDD268 pKa = 3.14MFEE271 pKa = 4.66RR272 pKa = 11.84NGRR275 pKa = 11.84VVEE278 pKa = 4.0RR279 pKa = 11.84RR280 pKa = 11.84GWPLASYY287 pKa = 10.37RR288 pKa = 11.84QFLRR292 pKa = 11.84QAAGMCRR299 pKa = 11.84GRR301 pKa = 11.84DD302 pKa = 3.12LRR304 pKa = 11.84ARR306 pKa = 11.84VEE308 pKa = 4.11LCLEE312 pKa = 4.03PLKK315 pKa = 11.02CRR317 pKa = 11.84VITKK321 pKa = 10.26GEE323 pKa = 4.07SVPYY327 pKa = 9.8FVAQTFQKK335 pKa = 10.96SMWKK339 pKa = 10.17ALQEE343 pKa = 3.93TGAFKK348 pKa = 9.95LTSCPVDD355 pKa = 3.13ASMLYY360 pKa = 10.64GIEE363 pKa = 5.22LSTKK367 pKa = 10.68DD368 pKa = 3.78LDD370 pKa = 4.34LPFDD374 pKa = 3.36QWVSGDD380 pKa = 3.54YY381 pKa = 10.74SAATDD386 pKa = 3.88GLTLEE391 pKa = 5.1VNQCCLRR398 pKa = 11.84AMLDD402 pKa = 3.48AFQATPEE409 pKa = 3.93EE410 pKa = 4.57RR411 pKa = 11.84EE412 pKa = 4.16VCSKK416 pKa = 11.09VLGCHH421 pKa = 4.83EE422 pKa = 3.96VSYY425 pKa = 10.69PDD427 pKa = 4.53RR428 pKa = 11.84LDD430 pKa = 3.78PEE432 pKa = 4.96ANGLEE437 pKa = 4.37PFHH440 pKa = 6.33MLNGQLMGSVLSFPVLCAVNLAAYY464 pKa = 7.54WCALEE469 pKa = 5.16EE470 pKa = 4.0YY471 pKa = 9.57TGRR474 pKa = 11.84KK475 pKa = 8.16FKK477 pKa = 10.96KK478 pKa = 8.37EE479 pKa = 3.8QLPVLVNGDD488 pKa = 4.35DD489 pKa = 3.95ILFKK493 pKa = 11.24ANAAFYY499 pKa = 9.0EE500 pKa = 4.68VWKK503 pKa = 10.24KK504 pKa = 6.7WVKK507 pKa = 10.22RR508 pKa = 11.84AGFSLSLGKK517 pKa = 10.4NYY519 pKa = 9.9ISPNFITVNSEE530 pKa = 3.46SWLHH534 pKa = 6.17KK535 pKa = 9.5GGSYY539 pKa = 9.82FRR541 pKa = 11.84KK542 pKa = 10.14LPFLNCGLLLQEE554 pKa = 4.42AAGPYY559 pKa = 9.19KK560 pKa = 10.78VPLRR564 pKa = 11.84AEE566 pKa = 3.87TAEE569 pKa = 4.15RR570 pKa = 11.84PLIPKK575 pKa = 7.98LQWIIDD581 pKa = 3.77NCNNPARR588 pKa = 11.84AFDD591 pKa = 4.64RR592 pKa = 11.84IKK594 pKa = 10.27HH595 pKa = 4.76HH596 pKa = 5.75WRR598 pKa = 11.84RR599 pKa = 11.84SINVHH604 pKa = 4.87TEE606 pKa = 4.04FGRR609 pKa = 11.84FNLCAPMEE617 pKa = 4.37LGGCGLKK624 pKa = 10.61LPEE627 pKa = 4.23AVRR630 pKa = 11.84PAVNFTAFQCLLAGRR645 pKa = 11.84SHH647 pKa = 6.45QSYY650 pKa = 11.11KK651 pKa = 10.72NLEE654 pKa = 4.05GTDD657 pKa = 2.95IRR659 pKa = 11.84DD660 pKa = 3.5RR661 pKa = 11.84PKK663 pKa = 10.49TGFEE667 pKa = 4.26RR668 pKa = 11.84ISVAEE673 pKa = 4.01VSKK676 pKa = 10.93SINPLPTEE684 pKa = 4.15SRR686 pKa = 11.84LGTAVLRR693 pKa = 11.84SPFEE697 pKa = 3.81PVRR700 pKa = 11.84GEE702 pKa = 3.94LEE704 pKa = 4.0VRR706 pKa = 11.84FEE708 pKa = 5.31DD709 pKa = 5.22PIACLRR715 pKa = 11.84VAGEE719 pKa = 4.38LNTAQTASDD728 pKa = 3.59QEE730 pKa = 4.23RR731 pKa = 11.84PVYY734 pKa = 9.61VLKK737 pKa = 10.74RR738 pKa = 11.84LSRR741 pKa = 11.84KK742 pKa = 9.47RR743 pKa = 11.84LGAVFSTGAKK753 pKa = 6.67ITKK756 pKa = 9.34PFLFPFEE763 pKa = 4.07VRR765 pKa = 11.84KK766 pKa = 9.6QLTRR770 pKa = 11.84GPTDD774 pKa = 3.77DD775 pKa = 4.7ASPPAMPEE783 pKa = 4.54DD784 pKa = 3.62IAEE787 pKa = 4.3MATEE791 pKa = 4.6LCKK794 pKa = 10.52SAPKK798 pKa = 9.65PSCGGGTSRR807 pKa = 11.84KK808 pKa = 6.6WW809 pKa = 3.06
Molecular weight: 91.35 kDa
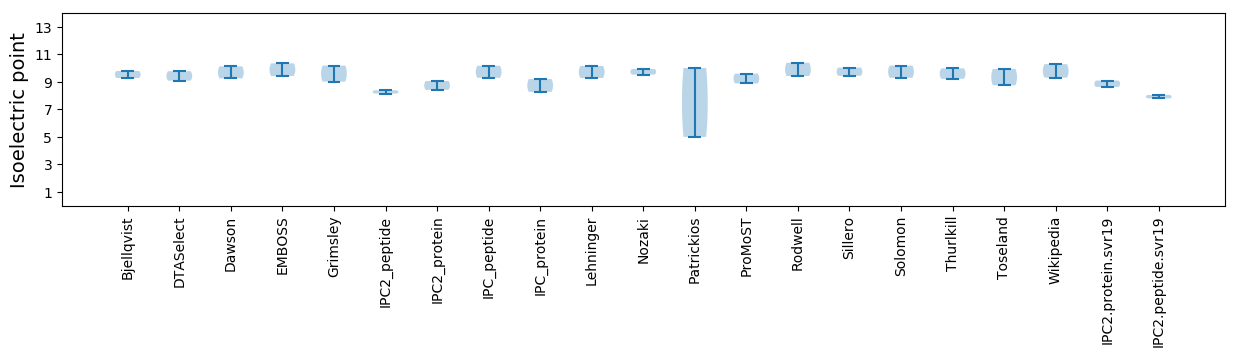
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIK3|A0A1L3KIK3_9VIRU Uncharacterized protein OS=Beihai narna-like virus 9 OX=1922461 PE=4 SV=1
MM1 pKa = 7.75AKK3 pKa = 10.1QNGVRR8 pKa = 11.84RR9 pKa = 11.84TVQKK13 pKa = 10.32KK14 pKa = 8.32VKK16 pKa = 9.58SDD18 pKa = 2.93RR19 pKa = 11.84RR20 pKa = 11.84SRR22 pKa = 11.84ADD24 pKa = 3.18VVQAQGTGRR33 pKa = 11.84TVQRR37 pKa = 11.84AFAGTKK43 pKa = 9.71RR44 pKa = 11.84ATGLEE49 pKa = 4.02GWDD52 pKa = 3.52AFSPAHH58 pKa = 6.28LPLPRR63 pKa = 11.84SVGPYY68 pKa = 9.14TVTKK72 pKa = 7.44TTALISTNDD81 pKa = 3.92RR82 pKa = 11.84INIIGTFAEE91 pKa = 4.44TDD93 pKa = 3.39TTGTKK98 pKa = 9.87LWTNVGLVKK107 pKa = 10.44SVNSALPISDD117 pKa = 3.87PANTFLHH124 pKa = 6.73TIPFPGNNITGSGLSATPAAVSVQIMNPNPLQTSSGIVAAAVCPTQLDD172 pKa = 3.67LRR174 pKa = 11.84GRR176 pKa = 11.84TEE178 pKa = 3.37TWAEE182 pKa = 3.84FGTEE186 pKa = 4.91FISYY190 pKa = 7.4MRR192 pKa = 11.84PRR194 pKa = 11.84LMSAGKK200 pKa = 9.4LALRR204 pKa = 11.84GVQMDD209 pKa = 4.49SYY211 pKa = 8.68PLNMGALAEE220 pKa = 4.02FRR222 pKa = 11.84PVNGFGDD229 pKa = 3.93TTIQWDD235 pKa = 4.14SANPTHH241 pKa = 6.19PQGWAPIVIVNQASSEE257 pKa = 4.03DD258 pKa = 3.87PPLEE262 pKa = 4.34LNLLVTIEE270 pKa = 3.43WRR272 pKa = 11.84TRR274 pKa = 11.84FDD276 pKa = 2.88IGNPAVSSHH285 pKa = 5.01SHH287 pKa = 6.36HH288 pKa = 6.42GVSSDD293 pKa = 3.47QHH295 pKa = 5.31WDD297 pKa = 3.16RR298 pKa = 11.84LIRR301 pKa = 11.84AATNRR306 pKa = 11.84GAGAMDD312 pKa = 3.0IVEE315 pKa = 4.34RR316 pKa = 11.84VANAGTAAIGAYY328 pKa = 9.46NAARR332 pKa = 11.84PILNQLPMLANFF344 pKa = 4.48
MM1 pKa = 7.75AKK3 pKa = 10.1QNGVRR8 pKa = 11.84RR9 pKa = 11.84TVQKK13 pKa = 10.32KK14 pKa = 8.32VKK16 pKa = 9.58SDD18 pKa = 2.93RR19 pKa = 11.84RR20 pKa = 11.84SRR22 pKa = 11.84ADD24 pKa = 3.18VVQAQGTGRR33 pKa = 11.84TVQRR37 pKa = 11.84AFAGTKK43 pKa = 9.71RR44 pKa = 11.84ATGLEE49 pKa = 4.02GWDD52 pKa = 3.52AFSPAHH58 pKa = 6.28LPLPRR63 pKa = 11.84SVGPYY68 pKa = 9.14TVTKK72 pKa = 7.44TTALISTNDD81 pKa = 3.92RR82 pKa = 11.84INIIGTFAEE91 pKa = 4.44TDD93 pKa = 3.39TTGTKK98 pKa = 9.87LWTNVGLVKK107 pKa = 10.44SVNSALPISDD117 pKa = 3.87PANTFLHH124 pKa = 6.73TIPFPGNNITGSGLSATPAAVSVQIMNPNPLQTSSGIVAAAVCPTQLDD172 pKa = 3.67LRR174 pKa = 11.84GRR176 pKa = 11.84TEE178 pKa = 3.37TWAEE182 pKa = 3.84FGTEE186 pKa = 4.91FISYY190 pKa = 7.4MRR192 pKa = 11.84PRR194 pKa = 11.84LMSAGKK200 pKa = 9.4LALRR204 pKa = 11.84GVQMDD209 pKa = 4.49SYY211 pKa = 8.68PLNMGALAEE220 pKa = 4.02FRR222 pKa = 11.84PVNGFGDD229 pKa = 3.93TTIQWDD235 pKa = 4.14SANPTHH241 pKa = 6.19PQGWAPIVIVNQASSEE257 pKa = 4.03DD258 pKa = 3.87PPLEE262 pKa = 4.34LNLLVTIEE270 pKa = 3.43WRR272 pKa = 11.84TRR274 pKa = 11.84FDD276 pKa = 2.88IGNPAVSSHH285 pKa = 5.01SHH287 pKa = 6.36HH288 pKa = 6.42GVSSDD293 pKa = 3.47QHH295 pKa = 5.31WDD297 pKa = 3.16RR298 pKa = 11.84LIRR301 pKa = 11.84AATNRR306 pKa = 11.84GAGAMDD312 pKa = 3.0IVEE315 pKa = 4.34RR316 pKa = 11.84VANAGTAAIGAYY328 pKa = 9.46NAARR332 pKa = 11.84PILNQLPMLANFF344 pKa = 4.48
Molecular weight: 36.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1153 |
344 |
809 |
576.5 |
64.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.714 ± 0.823 | 1.995 ± 0.864 |
4.163 ± 0.1 | 5.637 ± 1.384 |
4.683 ± 0.753 | 6.678 ± 0.741 |
2.082 ± 0.024 | 3.729 ± 0.909 |
4.683 ± 1.048 | 9.974 ± 1.372 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.995 ± 0.168 | 4.076 ± 1.028 |
6.852 ± 0.084 | 3.209 ± 0.436 |
7.892 ± 0.612 | 6.592 ± 0.195 |
5.637 ± 1.858 | 6.505 ± 0.239 |
2.255 ± 0.112 | 1.648 ± 0.246 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
