
Bovine faeces associated circular DNA molecule 1
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 5.09
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
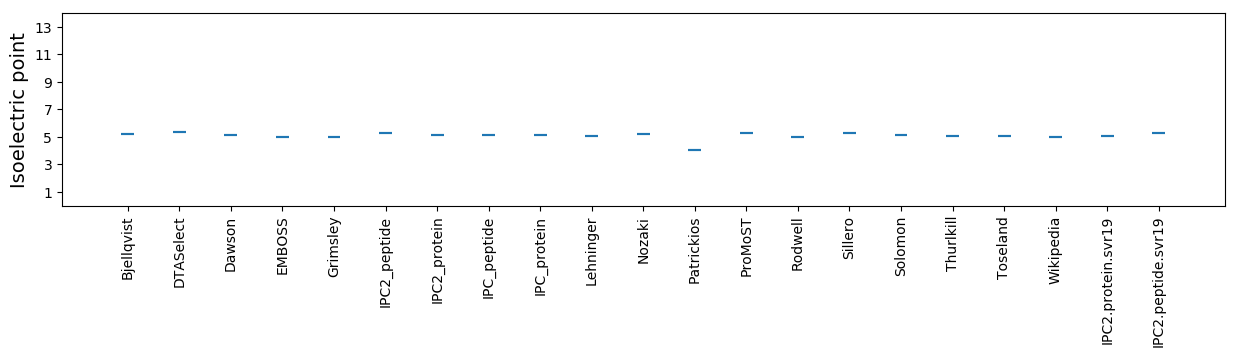
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A168MGH2|A0A168MGH2_9VIRU Replication associated protein OS=Bovine faeces associated circular DNA molecule 1 OX=1843763 PE=4 SV=1
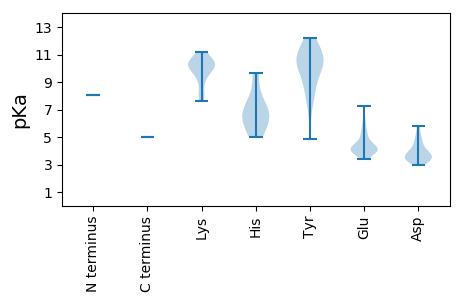
MM1 pKa = 8.05PSQQAKK7 pKa = 8.01VWTMTVKK14 pKa = 10.42NWDD17 pKa = 3.68LFDD20 pKa = 3.79NQEE23 pKa = 3.82WEE25 pKa = 4.3EE26 pKa = 4.18RR27 pKa = 11.84IQNQTEE33 pKa = 3.88PDD35 pKa = 3.22GSKK38 pKa = 10.07IIRR41 pKa = 11.84YY42 pKa = 9.33LSIGKK47 pKa = 8.08HH48 pKa = 5.01TGQQTGYY55 pKa = 7.19QHH57 pKa = 6.61CHH59 pKa = 5.67FNLEE63 pKa = 4.19LEE65 pKa = 4.6KK66 pKa = 10.52KK67 pKa = 8.93KK68 pKa = 9.56TMAWIKK74 pKa = 10.48RR75 pKa = 11.84EE76 pKa = 3.99LFNRR80 pKa = 11.84EE81 pKa = 4.55DD82 pKa = 3.18IHH84 pKa = 7.78CEE86 pKa = 3.38PRR88 pKa = 11.84RR89 pKa = 11.84GSRR92 pKa = 11.84EE93 pKa = 3.68QCDD96 pKa = 3.42SYY98 pKa = 11.54LNKK101 pKa = 10.79DD102 pKa = 3.51GEE104 pKa = 4.54FKK106 pKa = 11.14VIINTRR112 pKa = 11.84QIKK115 pKa = 9.24PGRR118 pKa = 11.84RR119 pKa = 11.84TDD121 pKa = 3.78LDD123 pKa = 5.18DD124 pKa = 3.31IHH126 pKa = 9.68DD127 pKa = 4.48MIKK130 pKa = 10.47EE131 pKa = 4.03GASLYY136 pKa = 10.74DD137 pKa = 3.58CYY139 pKa = 10.79EE140 pKa = 3.85EE141 pKa = 5.23HH142 pKa = 6.79FGTVVRR148 pKa = 11.84CEE150 pKa = 3.74RR151 pKa = 11.84GLRR154 pKa = 11.84DD155 pKa = 3.79YY156 pKa = 10.93IALRR160 pKa = 11.84DD161 pKa = 3.9TILATKK167 pKa = 9.79KK168 pKa = 10.11KK169 pKa = 9.13YY170 pKa = 8.43PAPEE174 pKa = 3.98VIVYY178 pKa = 8.34VGPSGSGKK186 pKa = 9.63SWHH189 pKa = 6.79CSEE192 pKa = 5.46DD193 pKa = 3.35PDD195 pKa = 4.39YY196 pKa = 11.81EE197 pKa = 4.09EE198 pKa = 4.18GGYY201 pKa = 10.19RR202 pKa = 11.84FSIQMDD208 pKa = 3.38SKK210 pKa = 11.09IYY212 pKa = 10.17FDD214 pKa = 4.78GYY216 pKa = 11.33NNQKK220 pKa = 7.65TLWFDD225 pKa = 3.45EE226 pKa = 4.91FSGKK230 pKa = 8.12TMPFTKK236 pKa = 10.15FCQIADD242 pKa = 4.56RR243 pKa = 11.84YY244 pKa = 8.04PGRR247 pKa = 11.84YY248 pKa = 4.83EE249 pKa = 4.02TKK251 pKa = 9.69GGSVLIYY258 pKa = 9.95GLKK261 pKa = 10.17KK262 pKa = 10.08ILISTVEE269 pKa = 4.28YY270 pKa = 9.13PALWWSGSDD279 pKa = 4.05RR280 pKa = 11.84YY281 pKa = 11.21NKK283 pKa = 10.46DD284 pKa = 2.94PEE286 pKa = 4.12QLFRR290 pKa = 11.84RR291 pKa = 11.84ITKK294 pKa = 10.02CYY296 pKa = 10.18YY297 pKa = 10.17LGQPRR302 pKa = 11.84IKK304 pKa = 10.16EE305 pKa = 4.36DD306 pKa = 3.26GDD308 pKa = 3.37IEE310 pKa = 4.22YY311 pKa = 10.37AIPLEE316 pKa = 4.1FNPRR320 pKa = 11.84HH321 pKa = 6.29LRR323 pKa = 11.84TQYY326 pKa = 11.05DD327 pKa = 3.74EE328 pKa = 5.75EE329 pKa = 4.16ILKK332 pKa = 11.16GNVKK336 pKa = 10.31YY337 pKa = 10.45PSDD340 pKa = 3.69LMEE343 pKa = 4.77EE344 pKa = 4.3EE345 pKa = 5.32PIATAEE351 pKa = 4.11EE352 pKa = 4.15AVKK355 pKa = 10.18EE356 pKa = 4.0LKK358 pKa = 10.45KK359 pKa = 10.83LLEE362 pKa = 3.92QSRR365 pKa = 11.84EE366 pKa = 3.84EE367 pKa = 5.55DD368 pKa = 3.25EE369 pKa = 4.12EE370 pKa = 4.45QPIKK374 pKa = 10.46RR375 pKa = 11.84RR376 pKa = 11.84RR377 pKa = 11.84SRR379 pKa = 11.84SRR381 pKa = 11.84SRR383 pKa = 11.84EE384 pKa = 3.75DD385 pKa = 4.9DD386 pKa = 3.41EE387 pKa = 7.24DD388 pKa = 3.84EE389 pKa = 6.41AEE391 pKa = 5.07DD392 pKa = 5.37DD393 pKa = 4.03YY394 pKa = 12.18CEE396 pKa = 6.14LIDD399 pKa = 5.82DD400 pKa = 4.11NN401 pKa = 5.01
MM1 pKa = 8.05PSQQAKK7 pKa = 8.01VWTMTVKK14 pKa = 10.42NWDD17 pKa = 3.68LFDD20 pKa = 3.79NQEE23 pKa = 3.82WEE25 pKa = 4.3EE26 pKa = 4.18RR27 pKa = 11.84IQNQTEE33 pKa = 3.88PDD35 pKa = 3.22GSKK38 pKa = 10.07IIRR41 pKa = 11.84YY42 pKa = 9.33LSIGKK47 pKa = 8.08HH48 pKa = 5.01TGQQTGYY55 pKa = 7.19QHH57 pKa = 6.61CHH59 pKa = 5.67FNLEE63 pKa = 4.19LEE65 pKa = 4.6KK66 pKa = 10.52KK67 pKa = 8.93KK68 pKa = 9.56TMAWIKK74 pKa = 10.48RR75 pKa = 11.84EE76 pKa = 3.99LFNRR80 pKa = 11.84EE81 pKa = 4.55DD82 pKa = 3.18IHH84 pKa = 7.78CEE86 pKa = 3.38PRR88 pKa = 11.84RR89 pKa = 11.84GSRR92 pKa = 11.84EE93 pKa = 3.68QCDD96 pKa = 3.42SYY98 pKa = 11.54LNKK101 pKa = 10.79DD102 pKa = 3.51GEE104 pKa = 4.54FKK106 pKa = 11.14VIINTRR112 pKa = 11.84QIKK115 pKa = 9.24PGRR118 pKa = 11.84RR119 pKa = 11.84TDD121 pKa = 3.78LDD123 pKa = 5.18DD124 pKa = 3.31IHH126 pKa = 9.68DD127 pKa = 4.48MIKK130 pKa = 10.47EE131 pKa = 4.03GASLYY136 pKa = 10.74DD137 pKa = 3.58CYY139 pKa = 10.79EE140 pKa = 3.85EE141 pKa = 5.23HH142 pKa = 6.79FGTVVRR148 pKa = 11.84CEE150 pKa = 3.74RR151 pKa = 11.84GLRR154 pKa = 11.84DD155 pKa = 3.79YY156 pKa = 10.93IALRR160 pKa = 11.84DD161 pKa = 3.9TILATKK167 pKa = 9.79KK168 pKa = 10.11KK169 pKa = 9.13YY170 pKa = 8.43PAPEE174 pKa = 3.98VIVYY178 pKa = 8.34VGPSGSGKK186 pKa = 9.63SWHH189 pKa = 6.79CSEE192 pKa = 5.46DD193 pKa = 3.35PDD195 pKa = 4.39YY196 pKa = 11.81EE197 pKa = 4.09EE198 pKa = 4.18GGYY201 pKa = 10.19RR202 pKa = 11.84FSIQMDD208 pKa = 3.38SKK210 pKa = 11.09IYY212 pKa = 10.17FDD214 pKa = 4.78GYY216 pKa = 11.33NNQKK220 pKa = 7.65TLWFDD225 pKa = 3.45EE226 pKa = 4.91FSGKK230 pKa = 8.12TMPFTKK236 pKa = 10.15FCQIADD242 pKa = 4.56RR243 pKa = 11.84YY244 pKa = 8.04PGRR247 pKa = 11.84YY248 pKa = 4.83EE249 pKa = 4.02TKK251 pKa = 9.69GGSVLIYY258 pKa = 9.95GLKK261 pKa = 10.17KK262 pKa = 10.08ILISTVEE269 pKa = 4.28YY270 pKa = 9.13PALWWSGSDD279 pKa = 4.05RR280 pKa = 11.84YY281 pKa = 11.21NKK283 pKa = 10.46DD284 pKa = 2.94PEE286 pKa = 4.12QLFRR290 pKa = 11.84RR291 pKa = 11.84ITKK294 pKa = 10.02CYY296 pKa = 10.18YY297 pKa = 10.17LGQPRR302 pKa = 11.84IKK304 pKa = 10.16EE305 pKa = 4.36DD306 pKa = 3.26GDD308 pKa = 3.37IEE310 pKa = 4.22YY311 pKa = 10.37AIPLEE316 pKa = 4.1FNPRR320 pKa = 11.84HH321 pKa = 6.29LRR323 pKa = 11.84TQYY326 pKa = 11.05DD327 pKa = 3.74EE328 pKa = 5.75EE329 pKa = 4.16ILKK332 pKa = 11.16GNVKK336 pKa = 10.31YY337 pKa = 10.45PSDD340 pKa = 3.69LMEE343 pKa = 4.77EE344 pKa = 4.3EE345 pKa = 5.32PIATAEE351 pKa = 4.11EE352 pKa = 4.15AVKK355 pKa = 10.18EE356 pKa = 4.0LKK358 pKa = 10.45KK359 pKa = 10.83LLEE362 pKa = 3.92QSRR365 pKa = 11.84EE366 pKa = 3.84EE367 pKa = 5.55DD368 pKa = 3.25EE369 pKa = 4.12EE370 pKa = 4.45QPIKK374 pKa = 10.46RR375 pKa = 11.84RR376 pKa = 11.84RR377 pKa = 11.84SRR379 pKa = 11.84SRR381 pKa = 11.84SRR383 pKa = 11.84EE384 pKa = 3.75DD385 pKa = 4.9DD386 pKa = 3.41EE387 pKa = 7.24DD388 pKa = 3.84EE389 pKa = 6.41AEE391 pKa = 5.07DD392 pKa = 5.37DD393 pKa = 4.03YY394 pKa = 12.18CEE396 pKa = 6.14LIDD399 pKa = 5.82DD400 pKa = 4.11NN401 pKa = 5.01
Molecular weight: 47.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A168MGH2|A0A168MGH2_9VIRU Replication associated protein OS=Bovine faeces associated circular DNA molecule 1 OX=1843763 PE=4 SV=1
MM1 pKa = 8.05PSQQAKK7 pKa = 8.01VWTMTVKK14 pKa = 10.42NWDD17 pKa = 3.68LFDD20 pKa = 3.79NQEE23 pKa = 3.82WEE25 pKa = 4.3EE26 pKa = 4.18RR27 pKa = 11.84IQNQTEE33 pKa = 3.88PDD35 pKa = 3.22GSKK38 pKa = 10.07IIRR41 pKa = 11.84YY42 pKa = 9.33LSIGKK47 pKa = 8.08HH48 pKa = 5.01TGQQTGYY55 pKa = 7.19QHH57 pKa = 6.61CHH59 pKa = 5.67FNLEE63 pKa = 4.19LEE65 pKa = 4.6KK66 pKa = 10.52KK67 pKa = 8.93KK68 pKa = 9.56TMAWIKK74 pKa = 10.48RR75 pKa = 11.84EE76 pKa = 3.99LFNRR80 pKa = 11.84EE81 pKa = 4.55DD82 pKa = 3.18IHH84 pKa = 7.78CEE86 pKa = 3.38PRR88 pKa = 11.84RR89 pKa = 11.84GSRR92 pKa = 11.84EE93 pKa = 3.68QCDD96 pKa = 3.42SYY98 pKa = 11.54LNKK101 pKa = 10.79DD102 pKa = 3.51GEE104 pKa = 4.54FKK106 pKa = 11.14VIINTRR112 pKa = 11.84QIKK115 pKa = 9.24PGRR118 pKa = 11.84RR119 pKa = 11.84TDD121 pKa = 3.78LDD123 pKa = 5.18DD124 pKa = 3.31IHH126 pKa = 9.68DD127 pKa = 4.48MIKK130 pKa = 10.47EE131 pKa = 4.03GASLYY136 pKa = 10.74DD137 pKa = 3.58CYY139 pKa = 10.79EE140 pKa = 3.85EE141 pKa = 5.23HH142 pKa = 6.79FGTVVRR148 pKa = 11.84CEE150 pKa = 3.74RR151 pKa = 11.84GLRR154 pKa = 11.84DD155 pKa = 3.79YY156 pKa = 10.93IALRR160 pKa = 11.84DD161 pKa = 3.9TILATKK167 pKa = 9.79KK168 pKa = 10.11KK169 pKa = 9.13YY170 pKa = 8.43PAPEE174 pKa = 3.98VIVYY178 pKa = 8.34VGPSGSGKK186 pKa = 9.63SWHH189 pKa = 6.79CSEE192 pKa = 5.46DD193 pKa = 3.35PDD195 pKa = 4.39YY196 pKa = 11.81EE197 pKa = 4.09EE198 pKa = 4.18GGYY201 pKa = 10.19RR202 pKa = 11.84FSIQMDD208 pKa = 3.38SKK210 pKa = 11.09IYY212 pKa = 10.17FDD214 pKa = 4.78GYY216 pKa = 11.33NNQKK220 pKa = 7.65TLWFDD225 pKa = 3.45EE226 pKa = 4.91FSGKK230 pKa = 8.12TMPFTKK236 pKa = 10.15FCQIADD242 pKa = 4.56RR243 pKa = 11.84YY244 pKa = 8.04PGRR247 pKa = 11.84YY248 pKa = 4.83EE249 pKa = 4.02TKK251 pKa = 9.69GGSVLIYY258 pKa = 9.95GLKK261 pKa = 10.17KK262 pKa = 10.08ILISTVEE269 pKa = 4.28YY270 pKa = 9.13PALWWSGSDD279 pKa = 4.05RR280 pKa = 11.84YY281 pKa = 11.21NKK283 pKa = 10.46DD284 pKa = 2.94PEE286 pKa = 4.12QLFRR290 pKa = 11.84RR291 pKa = 11.84ITKK294 pKa = 10.02CYY296 pKa = 10.18YY297 pKa = 10.17LGQPRR302 pKa = 11.84IKK304 pKa = 10.16EE305 pKa = 4.36DD306 pKa = 3.26GDD308 pKa = 3.37IEE310 pKa = 4.22YY311 pKa = 10.37AIPLEE316 pKa = 4.1FNPRR320 pKa = 11.84HH321 pKa = 6.29LRR323 pKa = 11.84TQYY326 pKa = 11.05DD327 pKa = 3.74EE328 pKa = 5.75EE329 pKa = 4.16ILKK332 pKa = 11.16GNVKK336 pKa = 10.31YY337 pKa = 10.45PSDD340 pKa = 3.69LMEE343 pKa = 4.77EE344 pKa = 4.3EE345 pKa = 5.32PIATAEE351 pKa = 4.11EE352 pKa = 4.15AVKK355 pKa = 10.18EE356 pKa = 4.0LKK358 pKa = 10.45KK359 pKa = 10.83LLEE362 pKa = 3.92QSRR365 pKa = 11.84EE366 pKa = 3.84EE367 pKa = 5.55DD368 pKa = 3.25EE369 pKa = 4.12EE370 pKa = 4.45QPIKK374 pKa = 10.46RR375 pKa = 11.84RR376 pKa = 11.84RR377 pKa = 11.84SRR379 pKa = 11.84SRR381 pKa = 11.84SRR383 pKa = 11.84EE384 pKa = 3.75DD385 pKa = 4.9DD386 pKa = 3.41EE387 pKa = 7.24DD388 pKa = 3.84EE389 pKa = 6.41AEE391 pKa = 5.07DD392 pKa = 5.37DD393 pKa = 4.03YY394 pKa = 12.18CEE396 pKa = 6.14LIDD399 pKa = 5.82DD400 pKa = 4.11NN401 pKa = 5.01
MM1 pKa = 8.05PSQQAKK7 pKa = 8.01VWTMTVKK14 pKa = 10.42NWDD17 pKa = 3.68LFDD20 pKa = 3.79NQEE23 pKa = 3.82WEE25 pKa = 4.3EE26 pKa = 4.18RR27 pKa = 11.84IQNQTEE33 pKa = 3.88PDD35 pKa = 3.22GSKK38 pKa = 10.07IIRR41 pKa = 11.84YY42 pKa = 9.33LSIGKK47 pKa = 8.08HH48 pKa = 5.01TGQQTGYY55 pKa = 7.19QHH57 pKa = 6.61CHH59 pKa = 5.67FNLEE63 pKa = 4.19LEE65 pKa = 4.6KK66 pKa = 10.52KK67 pKa = 8.93KK68 pKa = 9.56TMAWIKK74 pKa = 10.48RR75 pKa = 11.84EE76 pKa = 3.99LFNRR80 pKa = 11.84EE81 pKa = 4.55DD82 pKa = 3.18IHH84 pKa = 7.78CEE86 pKa = 3.38PRR88 pKa = 11.84RR89 pKa = 11.84GSRR92 pKa = 11.84EE93 pKa = 3.68QCDD96 pKa = 3.42SYY98 pKa = 11.54LNKK101 pKa = 10.79DD102 pKa = 3.51GEE104 pKa = 4.54FKK106 pKa = 11.14VIINTRR112 pKa = 11.84QIKK115 pKa = 9.24PGRR118 pKa = 11.84RR119 pKa = 11.84TDD121 pKa = 3.78LDD123 pKa = 5.18DD124 pKa = 3.31IHH126 pKa = 9.68DD127 pKa = 4.48MIKK130 pKa = 10.47EE131 pKa = 4.03GASLYY136 pKa = 10.74DD137 pKa = 3.58CYY139 pKa = 10.79EE140 pKa = 3.85EE141 pKa = 5.23HH142 pKa = 6.79FGTVVRR148 pKa = 11.84CEE150 pKa = 3.74RR151 pKa = 11.84GLRR154 pKa = 11.84DD155 pKa = 3.79YY156 pKa = 10.93IALRR160 pKa = 11.84DD161 pKa = 3.9TILATKK167 pKa = 9.79KK168 pKa = 10.11KK169 pKa = 9.13YY170 pKa = 8.43PAPEE174 pKa = 3.98VIVYY178 pKa = 8.34VGPSGSGKK186 pKa = 9.63SWHH189 pKa = 6.79CSEE192 pKa = 5.46DD193 pKa = 3.35PDD195 pKa = 4.39YY196 pKa = 11.81EE197 pKa = 4.09EE198 pKa = 4.18GGYY201 pKa = 10.19RR202 pKa = 11.84FSIQMDD208 pKa = 3.38SKK210 pKa = 11.09IYY212 pKa = 10.17FDD214 pKa = 4.78GYY216 pKa = 11.33NNQKK220 pKa = 7.65TLWFDD225 pKa = 3.45EE226 pKa = 4.91FSGKK230 pKa = 8.12TMPFTKK236 pKa = 10.15FCQIADD242 pKa = 4.56RR243 pKa = 11.84YY244 pKa = 8.04PGRR247 pKa = 11.84YY248 pKa = 4.83EE249 pKa = 4.02TKK251 pKa = 9.69GGSVLIYY258 pKa = 9.95GLKK261 pKa = 10.17KK262 pKa = 10.08ILISTVEE269 pKa = 4.28YY270 pKa = 9.13PALWWSGSDD279 pKa = 4.05RR280 pKa = 11.84YY281 pKa = 11.21NKK283 pKa = 10.46DD284 pKa = 2.94PEE286 pKa = 4.12QLFRR290 pKa = 11.84RR291 pKa = 11.84ITKK294 pKa = 10.02CYY296 pKa = 10.18YY297 pKa = 10.17LGQPRR302 pKa = 11.84IKK304 pKa = 10.16EE305 pKa = 4.36DD306 pKa = 3.26GDD308 pKa = 3.37IEE310 pKa = 4.22YY311 pKa = 10.37AIPLEE316 pKa = 4.1FNPRR320 pKa = 11.84HH321 pKa = 6.29LRR323 pKa = 11.84TQYY326 pKa = 11.05DD327 pKa = 3.74EE328 pKa = 5.75EE329 pKa = 4.16ILKK332 pKa = 11.16GNVKK336 pKa = 10.31YY337 pKa = 10.45PSDD340 pKa = 3.69LMEE343 pKa = 4.77EE344 pKa = 4.3EE345 pKa = 5.32PIATAEE351 pKa = 4.11EE352 pKa = 4.15AVKK355 pKa = 10.18EE356 pKa = 4.0LKK358 pKa = 10.45KK359 pKa = 10.83LLEE362 pKa = 3.92QSRR365 pKa = 11.84EE366 pKa = 3.84EE367 pKa = 5.55DD368 pKa = 3.25EE369 pKa = 4.12EE370 pKa = 4.45QPIKK374 pKa = 10.46RR375 pKa = 11.84RR376 pKa = 11.84RR377 pKa = 11.84SRR379 pKa = 11.84SRR381 pKa = 11.84SRR383 pKa = 11.84EE384 pKa = 3.75DD385 pKa = 4.9DD386 pKa = 3.41EE387 pKa = 7.24DD388 pKa = 3.84EE389 pKa = 6.41AEE391 pKa = 5.07DD392 pKa = 5.37DD393 pKa = 4.03YY394 pKa = 12.18CEE396 pKa = 6.14LIDD399 pKa = 5.82DD400 pKa = 4.11NN401 pKa = 5.01
Molecular weight: 47.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
401 |
401 |
401 |
401.0 |
47.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.242 ± 0.0 | 2.244 ± 0.0 |
8.229 ± 0.0 | 10.973 ± 0.0 |
3.242 ± 0.0 | 6.234 ± 0.0 |
1.995 ± 0.0 | 6.983 ± 0.0 |
7.98 ± 0.0 | 6.484 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.746 ± 0.0 | 3.242 ± 0.0 |
4.489 ± 0.0 | 4.489 ± 0.0 |
7.481 ± 0.0 | 5.486 ± 0.0 |
4.738 ± 0.0 | 2.993 ± 0.0 |
1.995 ± 0.0 | 5.736 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
