
Euphorbia caput-medusae latent virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Capulavirus
Average proteome isoelectric point is 7.27
Get precalculated fractions of proteins
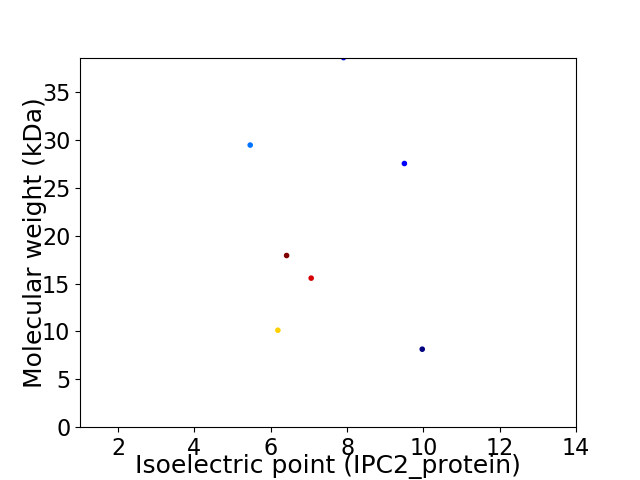
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
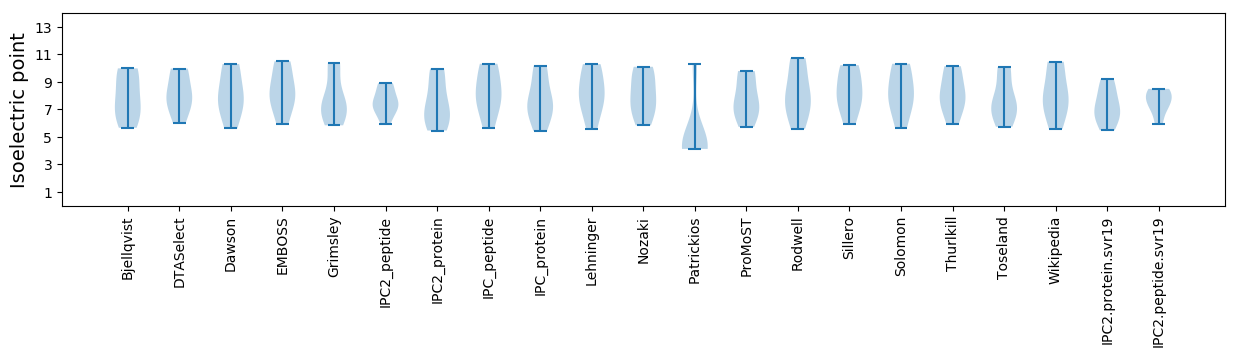
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A166V367|A0A166V367_9GEMI Replication-associated protein OS=Euphorbia caput-medusae latent virus OX=1853865 PE=3 SV=1
MM1 pKa = 7.67PRR3 pKa = 11.84QPNNTFRR10 pKa = 11.84LQGKK14 pKa = 9.52SIFLTYY20 pKa = 9.22PKK22 pKa = 10.43CPLLPIFLIDD32 pKa = 4.25YY33 pKa = 9.3LYY35 pKa = 11.06QLLNNFNPTYY45 pKa = 11.13ARR47 pKa = 11.84VCTEE51 pKa = 3.51NHH53 pKa = 6.07QDD55 pKa = 3.87GEE57 pKa = 4.4PHH59 pKa = 6.41LHH61 pKa = 6.89CLVQMDD67 pKa = 4.29KK68 pKa = 11.07RR69 pKa = 11.84LNTTNQRR76 pKa = 11.84FFDD79 pKa = 3.55IKK81 pKa = 10.94DD82 pKa = 3.56PNGVATYY89 pKa = 10.18HH90 pKa = 6.71PNCQVPRR97 pKa = 11.84RR98 pKa = 11.84DD99 pKa = 3.84ADD101 pKa = 3.58VADD104 pKa = 4.81YY105 pKa = 10.27IAKK108 pKa = 10.16GGQFEE113 pKa = 4.22EE114 pKa = 4.62RR115 pKa = 11.84GILRR119 pKa = 11.84ASRR122 pKa = 11.84RR123 pKa = 11.84SPKK126 pKa = 9.01KK127 pKa = 9.02TRR129 pKa = 11.84DD130 pKa = 3.88TIWASILTEE139 pKa = 4.11STSKK143 pKa = 11.01SEE145 pKa = 3.58FLARR149 pKa = 11.84VQHH152 pKa = 5.3EE153 pKa = 4.35QPYY156 pKa = 9.72VWATQLRR163 pKa = 11.84NLEE166 pKa = 4.0YY167 pKa = 10.4AANSKK172 pKa = 9.35WPEE175 pKa = 4.4PITVFEE181 pKa = 4.18PRR183 pKa = 11.84FVNFPRR189 pKa = 11.84IPEE192 pKa = 5.26PIQQWAEE199 pKa = 4.05TNLFTVSAHH208 pKa = 7.37AIQLLGADD216 pKa = 4.3LTIEE220 pKa = 4.8DD221 pKa = 4.55IRR223 pKa = 11.84WAHH226 pKa = 6.91DD227 pKa = 3.31LTTEE231 pKa = 4.99FINDD235 pKa = 3.43EE236 pKa = 4.72LINNIDD242 pKa = 3.62EE243 pKa = 5.0PIIINDD249 pKa = 4.03SGPEE253 pKa = 3.87TT254 pKa = 3.76
MM1 pKa = 7.67PRR3 pKa = 11.84QPNNTFRR10 pKa = 11.84LQGKK14 pKa = 9.52SIFLTYY20 pKa = 9.22PKK22 pKa = 10.43CPLLPIFLIDD32 pKa = 4.25YY33 pKa = 9.3LYY35 pKa = 11.06QLLNNFNPTYY45 pKa = 11.13ARR47 pKa = 11.84VCTEE51 pKa = 3.51NHH53 pKa = 6.07QDD55 pKa = 3.87GEE57 pKa = 4.4PHH59 pKa = 6.41LHH61 pKa = 6.89CLVQMDD67 pKa = 4.29KK68 pKa = 11.07RR69 pKa = 11.84LNTTNQRR76 pKa = 11.84FFDD79 pKa = 3.55IKK81 pKa = 10.94DD82 pKa = 3.56PNGVATYY89 pKa = 10.18HH90 pKa = 6.71PNCQVPRR97 pKa = 11.84RR98 pKa = 11.84DD99 pKa = 3.84ADD101 pKa = 3.58VADD104 pKa = 4.81YY105 pKa = 10.27IAKK108 pKa = 10.16GGQFEE113 pKa = 4.22EE114 pKa = 4.62RR115 pKa = 11.84GILRR119 pKa = 11.84ASRR122 pKa = 11.84RR123 pKa = 11.84SPKK126 pKa = 9.01KK127 pKa = 9.02TRR129 pKa = 11.84DD130 pKa = 3.88TIWASILTEE139 pKa = 4.11STSKK143 pKa = 11.01SEE145 pKa = 3.58FLARR149 pKa = 11.84VQHH152 pKa = 5.3EE153 pKa = 4.35QPYY156 pKa = 9.72VWATQLRR163 pKa = 11.84NLEE166 pKa = 4.0YY167 pKa = 10.4AANSKK172 pKa = 9.35WPEE175 pKa = 4.4PITVFEE181 pKa = 4.18PRR183 pKa = 11.84FVNFPRR189 pKa = 11.84IPEE192 pKa = 5.26PIQQWAEE199 pKa = 4.05TNLFTVSAHH208 pKa = 7.37AIQLLGADD216 pKa = 4.3LTIEE220 pKa = 4.8DD221 pKa = 4.55IRR223 pKa = 11.84WAHH226 pKa = 6.91DD227 pKa = 3.31LTTEE231 pKa = 4.99FINDD235 pKa = 3.43EE236 pKa = 4.72LINNIDD242 pKa = 3.62EE243 pKa = 5.0PIIINDD249 pKa = 4.03SGPEE253 pKa = 3.87TT254 pKa = 3.76
Molecular weight: 29.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A166V367|A0A166V367_9GEMI Replication-associated protein OS=Euphorbia caput-medusae latent virus OX=1853865 PE=3 SV=1
MM1 pKa = 6.44VTTRR5 pKa = 11.84SGRR8 pKa = 11.84NYY10 pKa = 9.72QALVPTWARR19 pKa = 11.84KK20 pKa = 9.04KK21 pKa = 10.39RR22 pKa = 11.84RR23 pKa = 11.84STLARR28 pKa = 11.84STIVGPIRR36 pKa = 11.84RR37 pKa = 11.84PNYY40 pKa = 9.27QYY42 pKa = 9.28KK43 pKa = 8.84TRR45 pKa = 11.84YY46 pKa = 6.85TPHH49 pKa = 7.09RR50 pKa = 11.84PQHH53 pKa = 6.26KK54 pKa = 9.36IHH56 pKa = 6.57SLSQTRR62 pKa = 11.84VVSGSDD68 pKa = 2.91NGYY71 pKa = 8.16GWHH74 pKa = 6.08VSGVNIGSGFEE85 pKa = 4.42DD86 pKa = 3.1RR87 pKa = 11.84HH88 pKa = 6.39SDD90 pKa = 3.36KK91 pKa = 11.09IKK93 pKa = 10.51IINLKK98 pKa = 10.72FMMQLKK104 pKa = 9.56TSDD107 pKa = 3.56AGQQASCYY115 pKa = 8.71HH116 pKa = 5.92NLYY119 pKa = 9.58MFLVKK124 pKa = 10.56DD125 pKa = 3.84NSGGAQVPKK134 pKa = 10.28FNSIVMMDD142 pKa = 3.75NSNPATAEE150 pKa = 3.69VDD152 pKa = 3.19HH153 pKa = 7.16DD154 pKa = 4.29SKK156 pKa = 11.8DD157 pKa = 2.86RR158 pKa = 11.84FTIVRR163 pKa = 11.84RR164 pKa = 11.84WKK166 pKa = 10.53FSFKK170 pKa = 10.92GNSSKK175 pKa = 11.26NGTAYY180 pKa = 10.55DD181 pKa = 3.8CARR184 pKa = 11.84NLIDD188 pKa = 3.8FNRR191 pKa = 11.84NVKK194 pKa = 9.89INSVSEE200 pKa = 4.35FKK202 pKa = 10.93SATDD206 pKa = 3.46GSYY209 pKa = 11.84ANTQKK214 pKa = 10.84NAWVLYY220 pKa = 9.29IVPQIYY226 pKa = 9.25DD227 pKa = 3.75CTVDD231 pKa = 3.17GHH233 pKa = 6.23VKK235 pKa = 9.79IKK237 pKa = 10.45YY238 pKa = 9.48VSIVV242 pKa = 2.95
MM1 pKa = 6.44VTTRR5 pKa = 11.84SGRR8 pKa = 11.84NYY10 pKa = 9.72QALVPTWARR19 pKa = 11.84KK20 pKa = 9.04KK21 pKa = 10.39RR22 pKa = 11.84RR23 pKa = 11.84STLARR28 pKa = 11.84STIVGPIRR36 pKa = 11.84RR37 pKa = 11.84PNYY40 pKa = 9.27QYY42 pKa = 9.28KK43 pKa = 8.84TRR45 pKa = 11.84YY46 pKa = 6.85TPHH49 pKa = 7.09RR50 pKa = 11.84PQHH53 pKa = 6.26KK54 pKa = 9.36IHH56 pKa = 6.57SLSQTRR62 pKa = 11.84VVSGSDD68 pKa = 2.91NGYY71 pKa = 8.16GWHH74 pKa = 6.08VSGVNIGSGFEE85 pKa = 4.42DD86 pKa = 3.1RR87 pKa = 11.84HH88 pKa = 6.39SDD90 pKa = 3.36KK91 pKa = 11.09IKK93 pKa = 10.51IINLKK98 pKa = 10.72FMMQLKK104 pKa = 9.56TSDD107 pKa = 3.56AGQQASCYY115 pKa = 8.71HH116 pKa = 5.92NLYY119 pKa = 9.58MFLVKK124 pKa = 10.56DD125 pKa = 3.84NSGGAQVPKK134 pKa = 10.28FNSIVMMDD142 pKa = 3.75NSNPATAEE150 pKa = 3.69VDD152 pKa = 3.19HH153 pKa = 7.16DD154 pKa = 4.29SKK156 pKa = 11.8DD157 pKa = 2.86RR158 pKa = 11.84FTIVRR163 pKa = 11.84RR164 pKa = 11.84WKK166 pKa = 10.53FSFKK170 pKa = 10.92GNSSKK175 pKa = 11.26NGTAYY180 pKa = 10.55DD181 pKa = 3.8CARR184 pKa = 11.84NLIDD188 pKa = 3.8FNRR191 pKa = 11.84NVKK194 pKa = 9.89INSVSEE200 pKa = 4.35FKK202 pKa = 10.93SATDD206 pKa = 3.46GSYY209 pKa = 11.84ANTQKK214 pKa = 10.84NAWVLYY220 pKa = 9.29IVPQIYY226 pKa = 9.25DD227 pKa = 3.75CTVDD231 pKa = 3.17GHH233 pKa = 6.23VKK235 pKa = 9.79IKK237 pKa = 10.45YY238 pKa = 9.48VSIVV242 pKa = 2.95
Molecular weight: 27.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1276 |
68 |
331 |
182.3 |
21.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.016 ± 0.532 | 1.489 ± 0.286 |
4.467 ± 0.601 | 5.329 ± 0.971 |
5.094 ± 0.524 | 5.408 ± 0.851 |
2.273 ± 0.28 | 6.505 ± 0.74 |
5.094 ± 0.922 | 7.837 ± 1.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.803 ± 0.474 | 6.426 ± 0.554 |
6.426 ± 1.007 | 5.329 ± 0.482 |
6.818 ± 0.233 | 7.21 ± 1.501 |
6.583 ± 0.444 | 4.389 ± 0.788 |
1.959 ± 0.115 | 4.545 ± 0.605 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
