
Citrus virus A
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Coguvirus; Coguvirus eburi
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
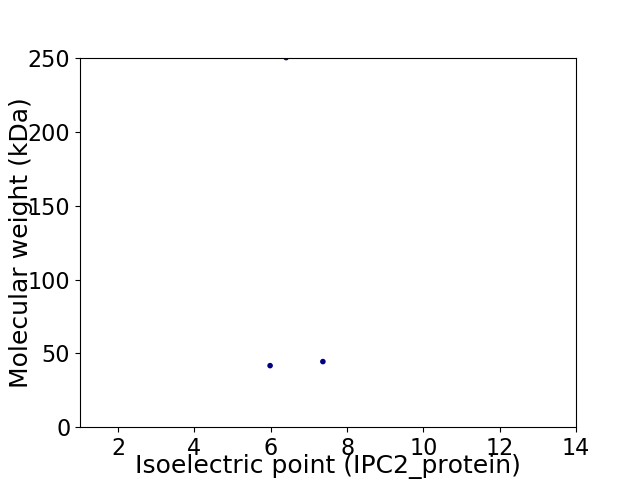
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3G2LX79|A0A3G2LX79_9VIRU Nucleocapsid protein OS=Citrus virus A OX=2484986 GN=s2gp2 PE=4 SV=1
MM1 pKa = 7.83ALHH4 pKa = 6.12QNPKK8 pKa = 9.28EE9 pKa = 3.97QSKK12 pKa = 10.74FDD14 pKa = 3.93ALFSQATSHH23 pKa = 6.52GEE25 pKa = 3.39EE26 pKa = 4.6HH27 pKa = 6.31YY28 pKa = 11.06RR29 pKa = 11.84EE30 pKa = 4.15FLTKK34 pKa = 10.39CSKK37 pKa = 10.22NLKK40 pKa = 9.41EE41 pKa = 4.19RR42 pKa = 11.84IKK44 pKa = 10.98RR45 pKa = 11.84EE46 pKa = 3.63VNARR50 pKa = 11.84KK51 pKa = 7.13MTSGGDD57 pKa = 3.46FLEE60 pKa = 4.77SFEE63 pKa = 4.2QAEE66 pKa = 4.44DD67 pKa = 3.69KK68 pKa = 11.03SGKK71 pKa = 9.77LDD73 pKa = 3.58NKK75 pKa = 10.32EE76 pKa = 4.09GEE78 pKa = 4.3LEE80 pKa = 4.03EE81 pKa = 5.84DD82 pKa = 3.2EE83 pKa = 6.07HH84 pKa = 8.65EE85 pKa = 4.96DD86 pKa = 3.46EE87 pKa = 4.33EE88 pKa = 5.19QEE90 pKa = 4.18VTEE93 pKa = 4.48GSGTKK98 pKa = 9.86KK99 pKa = 10.78DD100 pKa = 3.51IVVTDD105 pKa = 3.87AQLAAMWTEE114 pKa = 3.45IDD116 pKa = 4.28NIDD119 pKa = 3.33VSTFDD124 pKa = 3.68RR125 pKa = 11.84EE126 pKa = 4.29STKK129 pKa = 10.48VFDD132 pKa = 4.13YY133 pKa = 11.26QGFNPDD139 pKa = 2.93EE140 pKa = 4.27VLRR143 pKa = 11.84SLIINQRR150 pKa = 11.84KK151 pKa = 9.01NKK153 pKa = 9.7VSAADD158 pKa = 3.9FKK160 pKa = 11.69SDD162 pKa = 3.24ILLMCSLAIIKK173 pKa = 10.47GSINEE178 pKa = 4.18HH179 pKa = 5.25NFKK182 pKa = 10.71KK183 pKa = 10.67LSTEE187 pKa = 4.02GQSTVIRR194 pKa = 11.84LEE196 pKa = 3.52KK197 pKa = 10.08TYY199 pKa = 10.67GIKK202 pKa = 10.27RR203 pKa = 11.84GSGRR207 pKa = 11.84SEE209 pKa = 3.75PANVVTISRR218 pKa = 11.84IGATFPGKK226 pKa = 10.09IIQLLVEE233 pKa = 4.65NKK235 pKa = 9.44ASSRR239 pKa = 11.84KK240 pKa = 9.64FMGPLKK246 pKa = 10.67SHH248 pKa = 6.97TLPQFIRR255 pKa = 11.84HH256 pKa = 5.34QGFAAVIPTTLAEE269 pKa = 4.21STRR272 pKa = 11.84MFLLDD277 pKa = 3.28IVTAYY282 pKa = 10.73SVDD285 pKa = 3.47QSISISPNKK294 pKa = 9.38KK295 pKa = 9.8NKK297 pKa = 9.23PDD299 pKa = 3.57LASLYY304 pKa = 9.42EE305 pKa = 4.07KK306 pKa = 10.35QRR308 pKa = 11.84SFIQTTADD316 pKa = 2.86NKK318 pKa = 10.03YY319 pKa = 8.95PSEE322 pKa = 4.01IVRR325 pKa = 11.84KK326 pKa = 10.14KK327 pKa = 10.35IFTTIMIDD335 pKa = 3.46YY336 pKa = 10.42DD337 pKa = 3.97SLLTTARR344 pKa = 11.84ALLKK348 pKa = 9.94VIPDD352 pKa = 3.36FTIVTKK358 pKa = 10.76EE359 pKa = 3.92NFVSDD364 pKa = 3.6INSTHH369 pKa = 5.88II370 pKa = 3.9
MM1 pKa = 7.83ALHH4 pKa = 6.12QNPKK8 pKa = 9.28EE9 pKa = 3.97QSKK12 pKa = 10.74FDD14 pKa = 3.93ALFSQATSHH23 pKa = 6.52GEE25 pKa = 3.39EE26 pKa = 4.6HH27 pKa = 6.31YY28 pKa = 11.06RR29 pKa = 11.84EE30 pKa = 4.15FLTKK34 pKa = 10.39CSKK37 pKa = 10.22NLKK40 pKa = 9.41EE41 pKa = 4.19RR42 pKa = 11.84IKK44 pKa = 10.98RR45 pKa = 11.84EE46 pKa = 3.63VNARR50 pKa = 11.84KK51 pKa = 7.13MTSGGDD57 pKa = 3.46FLEE60 pKa = 4.77SFEE63 pKa = 4.2QAEE66 pKa = 4.44DD67 pKa = 3.69KK68 pKa = 11.03SGKK71 pKa = 9.77LDD73 pKa = 3.58NKK75 pKa = 10.32EE76 pKa = 4.09GEE78 pKa = 4.3LEE80 pKa = 4.03EE81 pKa = 5.84DD82 pKa = 3.2EE83 pKa = 6.07HH84 pKa = 8.65EE85 pKa = 4.96DD86 pKa = 3.46EE87 pKa = 4.33EE88 pKa = 5.19QEE90 pKa = 4.18VTEE93 pKa = 4.48GSGTKK98 pKa = 9.86KK99 pKa = 10.78DD100 pKa = 3.51IVVTDD105 pKa = 3.87AQLAAMWTEE114 pKa = 3.45IDD116 pKa = 4.28NIDD119 pKa = 3.33VSTFDD124 pKa = 3.68RR125 pKa = 11.84EE126 pKa = 4.29STKK129 pKa = 10.48VFDD132 pKa = 4.13YY133 pKa = 11.26QGFNPDD139 pKa = 2.93EE140 pKa = 4.27VLRR143 pKa = 11.84SLIINQRR150 pKa = 11.84KK151 pKa = 9.01NKK153 pKa = 9.7VSAADD158 pKa = 3.9FKK160 pKa = 11.69SDD162 pKa = 3.24ILLMCSLAIIKK173 pKa = 10.47GSINEE178 pKa = 4.18HH179 pKa = 5.25NFKK182 pKa = 10.71KK183 pKa = 10.67LSTEE187 pKa = 4.02GQSTVIRR194 pKa = 11.84LEE196 pKa = 3.52KK197 pKa = 10.08TYY199 pKa = 10.67GIKK202 pKa = 10.27RR203 pKa = 11.84GSGRR207 pKa = 11.84SEE209 pKa = 3.75PANVVTISRR218 pKa = 11.84IGATFPGKK226 pKa = 10.09IIQLLVEE233 pKa = 4.65NKK235 pKa = 9.44ASSRR239 pKa = 11.84KK240 pKa = 9.64FMGPLKK246 pKa = 10.67SHH248 pKa = 6.97TLPQFIRR255 pKa = 11.84HH256 pKa = 5.34QGFAAVIPTTLAEE269 pKa = 4.21STRR272 pKa = 11.84MFLLDD277 pKa = 3.28IVTAYY282 pKa = 10.73SVDD285 pKa = 3.47QSISISPNKK294 pKa = 9.38KK295 pKa = 9.8NKK297 pKa = 9.23PDD299 pKa = 3.57LASLYY304 pKa = 9.42EE305 pKa = 4.07KK306 pKa = 10.35QRR308 pKa = 11.84SFIQTTADD316 pKa = 2.86NKK318 pKa = 10.03YY319 pKa = 8.95PSEE322 pKa = 4.01IVRR325 pKa = 11.84KK326 pKa = 10.14KK327 pKa = 10.35IFTTIMIDD335 pKa = 3.46YY336 pKa = 10.42DD337 pKa = 3.97SLLTTARR344 pKa = 11.84ALLKK348 pKa = 9.94VIPDD352 pKa = 3.36FTIVTKK358 pKa = 10.76EE359 pKa = 3.92NFVSDD364 pKa = 3.6INSTHH369 pKa = 5.88II370 pKa = 3.9
Molecular weight: 41.73 kDa
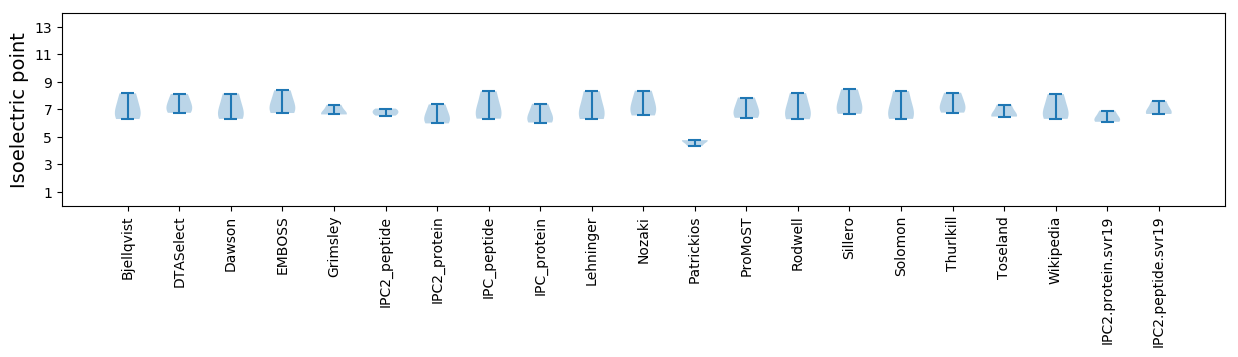
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G2LX79|A0A3G2LX79_9VIRU Nucleocapsid protein OS=Citrus virus A OX=2484986 GN=s2gp2 PE=4 SV=1
MM1 pKa = 7.14FKK3 pKa = 10.56KK4 pKa = 10.39VFRR7 pKa = 11.84RR8 pKa = 11.84ASGTPKK14 pKa = 10.21PVLQQATLQNRR25 pKa = 11.84PQRR28 pKa = 11.84QDD30 pKa = 2.47TDD32 pKa = 4.63SNVSRR37 pKa = 11.84AGKK40 pKa = 9.99NEE42 pKa = 3.65LVADD46 pKa = 4.77EE47 pKa = 4.5SLKK50 pKa = 9.97TADD53 pKa = 3.6RR54 pKa = 11.84NLYY57 pKa = 10.59KK58 pKa = 10.3FDD60 pKa = 4.75LPNNEE65 pKa = 4.21QGFDD69 pKa = 3.94DD70 pKa = 4.11LAKK73 pKa = 10.06EE74 pKa = 4.13YY75 pKa = 11.3SEE77 pKa = 4.17GTRR80 pKa = 11.84FQYY83 pKa = 10.77VALSGKK89 pKa = 10.1DD90 pKa = 3.41EE91 pKa = 4.16EE92 pKa = 5.13SYY94 pKa = 10.45SARR97 pKa = 11.84IRR99 pKa = 11.84RR100 pKa = 11.84LVTLTPRR107 pKa = 11.84MNKK110 pKa = 8.37DD111 pKa = 3.32TLPLTKK117 pKa = 9.59IPQLKK122 pKa = 10.17LIEE125 pKa = 4.54DD126 pKa = 4.25TEE128 pKa = 4.27QFNLSLAVPKK138 pKa = 10.34KK139 pKa = 8.27GKK141 pKa = 9.68PYY143 pKa = 10.03IRR145 pKa = 11.84LASVTAYY152 pKa = 9.89YY153 pKa = 10.3CPLVSSFSEE162 pKa = 4.32FTKK165 pKa = 10.91AGMSLHH171 pKa = 7.17DD172 pKa = 4.65SRR174 pKa = 11.84LSSDD178 pKa = 3.39TSVQSVTFNSNITQKK193 pKa = 11.23LEE195 pKa = 3.83LSLDD199 pKa = 3.47YY200 pKa = 10.88CIPRR204 pKa = 11.84SSASKK209 pKa = 8.53ITLNIARR216 pKa = 11.84EE217 pKa = 4.02QKK219 pKa = 9.71FLKK222 pKa = 10.56EE223 pKa = 3.98GEE225 pKa = 4.02EE226 pKa = 4.0WGAVQLLIKK235 pKa = 10.57LEE237 pKa = 4.24EE238 pKa = 4.31SDD240 pKa = 6.05FPYY243 pKa = 10.7CSNLKK248 pKa = 9.96EE249 pKa = 4.16ATAVMALPPAVLDD262 pKa = 3.72SHH264 pKa = 6.57NVNPNHH270 pKa = 7.06IDD272 pKa = 3.29TTIVEE277 pKa = 4.3TQRR280 pKa = 11.84RR281 pKa = 11.84KK282 pKa = 9.91VRR284 pKa = 11.84EE285 pKa = 3.67IYY287 pKa = 10.73EE288 pKa = 4.2NGDD291 pKa = 2.74ISNEE295 pKa = 3.86NEE297 pKa = 4.46PIKK300 pKa = 10.98EE301 pKa = 3.95MLGKK305 pKa = 10.35VSYY308 pKa = 10.33AKK310 pKa = 10.48SSISAKK316 pKa = 10.59VKK318 pKa = 9.66GQKK321 pKa = 7.78TNKK324 pKa = 9.71KK325 pKa = 9.22FGKK328 pKa = 9.36GWEE331 pKa = 4.01HH332 pKa = 6.12MEE334 pKa = 3.8FRR336 pKa = 11.84FPKK339 pKa = 8.45ITKK342 pKa = 9.1DD343 pKa = 3.45QEE345 pKa = 4.3SEE347 pKa = 4.08EE348 pKa = 4.31PEE350 pKa = 3.97EE351 pKa = 4.98DD352 pKa = 3.57GSIATPSLGSGRR364 pKa = 11.84NSVQEE369 pKa = 3.79IEE371 pKa = 4.48EE372 pKa = 4.23VVGTSEE378 pKa = 4.08TKK380 pKa = 10.23KK381 pKa = 10.0VNFNKK386 pKa = 10.08GVKK389 pKa = 9.8FNEE392 pKa = 4.37FGVV395 pKa = 3.64
MM1 pKa = 7.14FKK3 pKa = 10.56KK4 pKa = 10.39VFRR7 pKa = 11.84RR8 pKa = 11.84ASGTPKK14 pKa = 10.21PVLQQATLQNRR25 pKa = 11.84PQRR28 pKa = 11.84QDD30 pKa = 2.47TDD32 pKa = 4.63SNVSRR37 pKa = 11.84AGKK40 pKa = 9.99NEE42 pKa = 3.65LVADD46 pKa = 4.77EE47 pKa = 4.5SLKK50 pKa = 9.97TADD53 pKa = 3.6RR54 pKa = 11.84NLYY57 pKa = 10.59KK58 pKa = 10.3FDD60 pKa = 4.75LPNNEE65 pKa = 4.21QGFDD69 pKa = 3.94DD70 pKa = 4.11LAKK73 pKa = 10.06EE74 pKa = 4.13YY75 pKa = 11.3SEE77 pKa = 4.17GTRR80 pKa = 11.84FQYY83 pKa = 10.77VALSGKK89 pKa = 10.1DD90 pKa = 3.41EE91 pKa = 4.16EE92 pKa = 5.13SYY94 pKa = 10.45SARR97 pKa = 11.84IRR99 pKa = 11.84RR100 pKa = 11.84LVTLTPRR107 pKa = 11.84MNKK110 pKa = 8.37DD111 pKa = 3.32TLPLTKK117 pKa = 9.59IPQLKK122 pKa = 10.17LIEE125 pKa = 4.54DD126 pKa = 4.25TEE128 pKa = 4.27QFNLSLAVPKK138 pKa = 10.34KK139 pKa = 8.27GKK141 pKa = 9.68PYY143 pKa = 10.03IRR145 pKa = 11.84LASVTAYY152 pKa = 9.89YY153 pKa = 10.3CPLVSSFSEE162 pKa = 4.32FTKK165 pKa = 10.91AGMSLHH171 pKa = 7.17DD172 pKa = 4.65SRR174 pKa = 11.84LSSDD178 pKa = 3.39TSVQSVTFNSNITQKK193 pKa = 11.23LEE195 pKa = 3.83LSLDD199 pKa = 3.47YY200 pKa = 10.88CIPRR204 pKa = 11.84SSASKK209 pKa = 8.53ITLNIARR216 pKa = 11.84EE217 pKa = 4.02QKK219 pKa = 9.71FLKK222 pKa = 10.56EE223 pKa = 3.98GEE225 pKa = 4.02EE226 pKa = 4.0WGAVQLLIKK235 pKa = 10.57LEE237 pKa = 4.24EE238 pKa = 4.31SDD240 pKa = 6.05FPYY243 pKa = 10.7CSNLKK248 pKa = 9.96EE249 pKa = 4.16ATAVMALPPAVLDD262 pKa = 3.72SHH264 pKa = 6.57NVNPNHH270 pKa = 7.06IDD272 pKa = 3.29TTIVEE277 pKa = 4.3TQRR280 pKa = 11.84RR281 pKa = 11.84KK282 pKa = 9.91VRR284 pKa = 11.84EE285 pKa = 3.67IYY287 pKa = 10.73EE288 pKa = 4.2NGDD291 pKa = 2.74ISNEE295 pKa = 3.86NEE297 pKa = 4.46PIKK300 pKa = 10.98EE301 pKa = 3.95MLGKK305 pKa = 10.35VSYY308 pKa = 10.33AKK310 pKa = 10.48SSISAKK316 pKa = 10.59VKK318 pKa = 9.66GQKK321 pKa = 7.78TNKK324 pKa = 9.71KK325 pKa = 9.22FGKK328 pKa = 9.36GWEE331 pKa = 4.01HH332 pKa = 6.12MEE334 pKa = 3.8FRR336 pKa = 11.84FPKK339 pKa = 8.45ITKK342 pKa = 9.1DD343 pKa = 3.45QEE345 pKa = 4.3SEE347 pKa = 4.08EE348 pKa = 4.31PEE350 pKa = 3.97EE351 pKa = 4.98DD352 pKa = 3.57GSIATPSLGSGRR364 pKa = 11.84NSVQEE369 pKa = 3.79IEE371 pKa = 4.48EE372 pKa = 4.23VVGTSEE378 pKa = 4.08TKK380 pKa = 10.23KK381 pKa = 10.0VNFNKK386 pKa = 10.08GVKK389 pKa = 9.8FNEE392 pKa = 4.37FGVV395 pKa = 3.64
Molecular weight: 44.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2949 |
370 |
2184 |
983.0 |
112.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.68 ± 0.562 | 1.492 ± 0.439 |
5.866 ± 0.352 | 7.765 ± 0.555 |
4.747 ± 0.165 | 4.646 ± 0.142 |
1.831 ± 0.273 | 7.426 ± 0.961 |
8.647 ± 0.339 | 9.257 ± 0.678 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.34 ± 0.352 | 4.51 ± 0.466 |
3.594 ± 0.41 | 3.357 ± 0.442 |
4.781 ± 0.098 | 9.19 ± 0.15 |
5.866 ± 0.586 | 5.561 ± 0.261 |
1.051 ± 0.346 | 3.391 ± 0.557 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
