
Trichomonas vaginalis virus 3
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Trichomonasvirus
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q8V616|Q8V616_9VIRU Capsid protein OS=Trichomonas vaginalis virus 3 OX=170965 GN=cap PE=4 SV=1
MM1 pKa = 7.71DD2 pKa = 4.73HH3 pKa = 6.88VSDD6 pKa = 3.39IAYY9 pKa = 10.5LNFLRR14 pKa = 11.84WVLLPYY20 pKa = 10.33NGQTLRR26 pKa = 11.84PHH28 pKa = 6.37PSVWRR33 pKa = 11.84QTPYY37 pKa = 10.41PEE39 pKa = 4.17HH40 pKa = 6.9VNLKK44 pKa = 9.82FLNKK48 pKa = 9.83EE49 pKa = 3.83MEE51 pKa = 4.46LEE53 pKa = 4.05LFPLKK58 pKa = 10.34KK59 pKa = 10.48APQADD64 pKa = 3.8LKK66 pKa = 10.02VNCYY70 pKa = 9.75ARR72 pKa = 11.84NVLASTEE79 pKa = 4.1LTDD82 pKa = 5.96DD83 pKa = 4.45LLKK86 pKa = 10.78QSLPIGLNNDD96 pKa = 3.75SVCGIVIVLEE106 pKa = 4.0LLRR109 pKa = 11.84IAGVPSKK116 pKa = 10.22LLPIIGQAIANKK128 pKa = 10.45DD129 pKa = 3.56PFIKK133 pKa = 10.35EE134 pKa = 3.7LSDD137 pKa = 3.55FNKK140 pKa = 10.19MIGATTSRR148 pKa = 11.84IANILTEE155 pKa = 4.37CNTLIGRR162 pKa = 11.84GVKK165 pKa = 10.35SSDD168 pKa = 3.13PSADD172 pKa = 3.1LYY174 pKa = 11.37HH175 pKa = 6.94RR176 pKa = 11.84VAPEE180 pKa = 4.07GNRR183 pKa = 11.84HH184 pKa = 4.74EE185 pKa = 4.79AKK187 pKa = 10.1IPRR190 pKa = 11.84HH191 pKa = 5.28ILIEE195 pKa = 4.73AINKK199 pKa = 9.11IYY201 pKa = 10.19KK202 pKa = 10.2NEE204 pKa = 4.04MTDD207 pKa = 3.39MPPPGDD213 pKa = 3.73FKK215 pKa = 11.52LHH217 pKa = 7.23LITSPLWCKK226 pKa = 10.39AGSHH230 pKa = 5.76HH231 pKa = 6.8HH232 pKa = 5.98PHH234 pKa = 5.93FAKK237 pKa = 10.6YY238 pKa = 10.19SSRR241 pKa = 11.84LDD243 pKa = 3.54FVMDD247 pKa = 3.83VPADD251 pKa = 4.04KK252 pKa = 10.51IAAVPPSVFITQAEE266 pKa = 4.22KK267 pKa = 11.08LEE269 pKa = 4.45HH270 pKa = 6.31GKK272 pKa = 7.86TRR274 pKa = 11.84YY275 pKa = 9.27IYY277 pKa = 10.73NCDD280 pKa = 2.94TXSYY284 pKa = 10.86LFFDD288 pKa = 4.72YY289 pKa = 10.4ILHH292 pKa = 5.57YY293 pKa = 9.68VEE295 pKa = 5.77CVWSNEE301 pKa = 4.08SVLLNPAAMSVEE313 pKa = 4.15RR314 pKa = 11.84FSVLDD319 pKa = 3.52YY320 pKa = 11.16PEE322 pKa = 4.68YY323 pKa = 11.39CMIDD327 pKa = 3.35YY328 pKa = 10.75TDD330 pKa = 4.64FNSQHH335 pKa = 6.02SLEE338 pKa = 4.24SQKK341 pKa = 11.06LVFEE345 pKa = 4.52CLRR348 pKa = 11.84PYY350 pKa = 10.46LPSEE354 pKa = 3.78MHH356 pKa = 7.5PILDD360 pKa = 3.53WCIASMDD367 pKa = 3.45HH368 pKa = 6.37MEE370 pKa = 5.08IGGQHH375 pKa = 6.29WLSTLPSGHH384 pKa = 7.24RR385 pKa = 11.84ATTFINSVLNKK396 pKa = 10.14AYY398 pKa = 10.37LIPYY402 pKa = 9.55IGDD405 pKa = 3.36TTSFHH410 pKa = 6.98CGDD413 pKa = 4.64DD414 pKa = 3.49VLLCGKK420 pKa = 10.09YY421 pKa = 10.12DD422 pKa = 3.77YY423 pKa = 9.52QTLIDD428 pKa = 3.92TLPYY432 pKa = 10.15EE433 pKa = 4.39LNKK436 pKa = 10.4SKK438 pKa = 11.0QSFGPNAEE446 pKa = 3.89FLRR449 pKa = 11.84LHH451 pKa = 6.49RR452 pKa = 11.84RR453 pKa = 11.84AGDD456 pKa = 3.48VIGYY460 pKa = 7.13PSRR463 pKa = 11.84AVSSLVSGNWLSKK476 pKa = 9.65TSWEE480 pKa = 4.11WQPSLISVTNQCNVIISRR498 pKa = 11.84SQLNIRR504 pKa = 11.84FIPAMQQEE512 pKa = 4.32LRR514 pKa = 11.84NRR516 pKa = 11.84YY517 pKa = 7.74IDD519 pKa = 3.86KK520 pKa = 10.07MSEE523 pKa = 4.02PFDD526 pKa = 3.8VGSDD530 pKa = 3.98YY531 pKa = 11.52YY532 pKa = 11.78VMPGCPCYY540 pKa = 10.7SDD542 pKa = 3.74AATTIVPNVPQLEE555 pKa = 4.38CSDD558 pKa = 4.0VPFSQAQKK566 pKa = 10.74VFDD569 pKa = 4.09TMRR572 pKa = 11.84DD573 pKa = 2.99ICPEE577 pKa = 3.87FTTVNDD583 pKa = 3.63VIDD586 pKa = 3.74RR587 pKa = 11.84VLARR591 pKa = 11.84RR592 pKa = 11.84TSNAVKK598 pKa = 10.39NITYY602 pKa = 9.33NVCAPVAPQVCIAVNPAHH620 pKa = 5.92YY621 pKa = 10.32QFLLRR626 pKa = 11.84KK627 pKa = 8.91KK628 pKa = 9.1YY629 pKa = 10.5YY630 pKa = 8.88PRR632 pKa = 11.84EE633 pKa = 4.34HH634 pKa = 6.86IAPPGFDD641 pKa = 4.46DD642 pKa = 3.78STNSKK647 pKa = 10.55LVFSTYY653 pKa = 11.3DD654 pKa = 3.65LAPSIAMKK662 pKa = 10.29SCAVLTPAKK671 pKa = 9.59IICGHH676 pKa = 5.96GLRR679 pKa = 11.84SGG681 pKa = 3.43
MM1 pKa = 7.71DD2 pKa = 4.73HH3 pKa = 6.88VSDD6 pKa = 3.39IAYY9 pKa = 10.5LNFLRR14 pKa = 11.84WVLLPYY20 pKa = 10.33NGQTLRR26 pKa = 11.84PHH28 pKa = 6.37PSVWRR33 pKa = 11.84QTPYY37 pKa = 10.41PEE39 pKa = 4.17HH40 pKa = 6.9VNLKK44 pKa = 9.82FLNKK48 pKa = 9.83EE49 pKa = 3.83MEE51 pKa = 4.46LEE53 pKa = 4.05LFPLKK58 pKa = 10.34KK59 pKa = 10.48APQADD64 pKa = 3.8LKK66 pKa = 10.02VNCYY70 pKa = 9.75ARR72 pKa = 11.84NVLASTEE79 pKa = 4.1LTDD82 pKa = 5.96DD83 pKa = 4.45LLKK86 pKa = 10.78QSLPIGLNNDD96 pKa = 3.75SVCGIVIVLEE106 pKa = 4.0LLRR109 pKa = 11.84IAGVPSKK116 pKa = 10.22LLPIIGQAIANKK128 pKa = 10.45DD129 pKa = 3.56PFIKK133 pKa = 10.35EE134 pKa = 3.7LSDD137 pKa = 3.55FNKK140 pKa = 10.19MIGATTSRR148 pKa = 11.84IANILTEE155 pKa = 4.37CNTLIGRR162 pKa = 11.84GVKK165 pKa = 10.35SSDD168 pKa = 3.13PSADD172 pKa = 3.1LYY174 pKa = 11.37HH175 pKa = 6.94RR176 pKa = 11.84VAPEE180 pKa = 4.07GNRR183 pKa = 11.84HH184 pKa = 4.74EE185 pKa = 4.79AKK187 pKa = 10.1IPRR190 pKa = 11.84HH191 pKa = 5.28ILIEE195 pKa = 4.73AINKK199 pKa = 9.11IYY201 pKa = 10.19KK202 pKa = 10.2NEE204 pKa = 4.04MTDD207 pKa = 3.39MPPPGDD213 pKa = 3.73FKK215 pKa = 11.52LHH217 pKa = 7.23LITSPLWCKK226 pKa = 10.39AGSHH230 pKa = 5.76HH231 pKa = 6.8HH232 pKa = 5.98PHH234 pKa = 5.93FAKK237 pKa = 10.6YY238 pKa = 10.19SSRR241 pKa = 11.84LDD243 pKa = 3.54FVMDD247 pKa = 3.83VPADD251 pKa = 4.04KK252 pKa = 10.51IAAVPPSVFITQAEE266 pKa = 4.22KK267 pKa = 11.08LEE269 pKa = 4.45HH270 pKa = 6.31GKK272 pKa = 7.86TRR274 pKa = 11.84YY275 pKa = 9.27IYY277 pKa = 10.73NCDD280 pKa = 2.94TXSYY284 pKa = 10.86LFFDD288 pKa = 4.72YY289 pKa = 10.4ILHH292 pKa = 5.57YY293 pKa = 9.68VEE295 pKa = 5.77CVWSNEE301 pKa = 4.08SVLLNPAAMSVEE313 pKa = 4.15RR314 pKa = 11.84FSVLDD319 pKa = 3.52YY320 pKa = 11.16PEE322 pKa = 4.68YY323 pKa = 11.39CMIDD327 pKa = 3.35YY328 pKa = 10.75TDD330 pKa = 4.64FNSQHH335 pKa = 6.02SLEE338 pKa = 4.24SQKK341 pKa = 11.06LVFEE345 pKa = 4.52CLRR348 pKa = 11.84PYY350 pKa = 10.46LPSEE354 pKa = 3.78MHH356 pKa = 7.5PILDD360 pKa = 3.53WCIASMDD367 pKa = 3.45HH368 pKa = 6.37MEE370 pKa = 5.08IGGQHH375 pKa = 6.29WLSTLPSGHH384 pKa = 7.24RR385 pKa = 11.84ATTFINSVLNKK396 pKa = 10.14AYY398 pKa = 10.37LIPYY402 pKa = 9.55IGDD405 pKa = 3.36TTSFHH410 pKa = 6.98CGDD413 pKa = 4.64DD414 pKa = 3.49VLLCGKK420 pKa = 10.09YY421 pKa = 10.12DD422 pKa = 3.77YY423 pKa = 9.52QTLIDD428 pKa = 3.92TLPYY432 pKa = 10.15EE433 pKa = 4.39LNKK436 pKa = 10.4SKK438 pKa = 11.0QSFGPNAEE446 pKa = 3.89FLRR449 pKa = 11.84LHH451 pKa = 6.49RR452 pKa = 11.84RR453 pKa = 11.84AGDD456 pKa = 3.48VIGYY460 pKa = 7.13PSRR463 pKa = 11.84AVSSLVSGNWLSKK476 pKa = 9.65TSWEE480 pKa = 4.11WQPSLISVTNQCNVIISRR498 pKa = 11.84SQLNIRR504 pKa = 11.84FIPAMQQEE512 pKa = 4.32LRR514 pKa = 11.84NRR516 pKa = 11.84YY517 pKa = 7.74IDD519 pKa = 3.86KK520 pKa = 10.07MSEE523 pKa = 4.02PFDD526 pKa = 3.8VGSDD530 pKa = 3.98YY531 pKa = 11.52YY532 pKa = 11.78VMPGCPCYY540 pKa = 10.7SDD542 pKa = 3.74AATTIVPNVPQLEE555 pKa = 4.38CSDD558 pKa = 4.0VPFSQAQKK566 pKa = 10.74VFDD569 pKa = 4.09TMRR572 pKa = 11.84DD573 pKa = 2.99ICPEE577 pKa = 3.87FTTVNDD583 pKa = 3.63VIDD586 pKa = 3.74RR587 pKa = 11.84VLARR591 pKa = 11.84RR592 pKa = 11.84TSNAVKK598 pKa = 10.39NITYY602 pKa = 9.33NVCAPVAPQVCIAVNPAHH620 pKa = 5.92YY621 pKa = 10.32QFLLRR626 pKa = 11.84KK627 pKa = 8.91KK628 pKa = 9.1YY629 pKa = 10.5YY630 pKa = 8.88PRR632 pKa = 11.84EE633 pKa = 4.34HH634 pKa = 6.86IAPPGFDD641 pKa = 4.46DD642 pKa = 3.78STNSKK647 pKa = 10.55LVFSTYY653 pKa = 11.3DD654 pKa = 3.65LAPSIAMKK662 pKa = 10.29SCAVLTPAKK671 pKa = 9.59IICGHH676 pKa = 5.96GLRR679 pKa = 11.84SGG681 pKa = 3.43
Molecular weight: 76.8 kDa
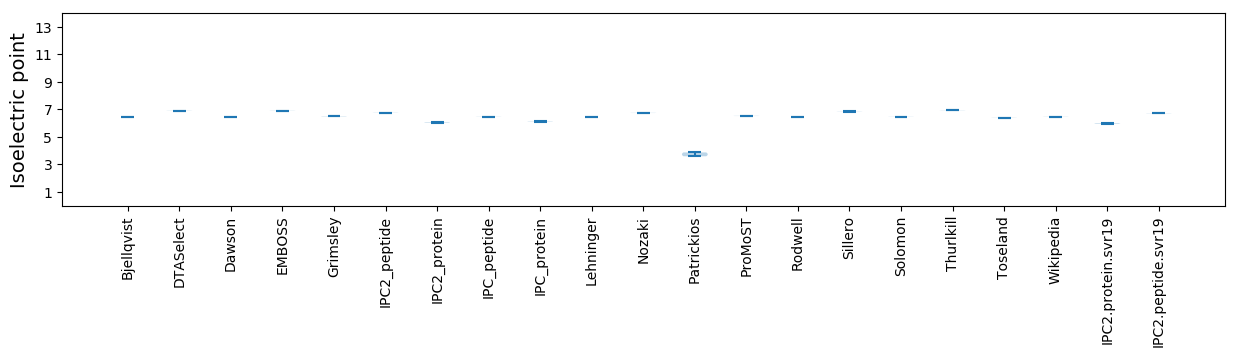
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8V616|Q8V616_9VIRU Capsid protein OS=Trichomonas vaginalis virus 3 OX=170965 GN=cap PE=4 SV=1
MM1 pKa = 7.64SAPEE5 pKa = 3.95PLNTEE10 pKa = 3.76VRR12 pKa = 11.84SPNGVSEE19 pKa = 5.13AIEE22 pKa = 4.22TQNMAVTQSSVSNEE36 pKa = 3.81IKK38 pKa = 10.69NDD40 pKa = 3.7TQSDD44 pKa = 4.17LQTLKK49 pKa = 10.89KK50 pKa = 10.1QLQPLYY56 pKa = 10.49RR57 pKa = 11.84STDD60 pKa = 3.42FNTLYY65 pKa = 11.07NFFYY69 pKa = 10.84GLDD72 pKa = 3.68VPASTDD78 pKa = 3.36RR79 pKa = 11.84VGHH82 pKa = 6.72AIQRR86 pKa = 11.84NTSVNDD92 pKa = 3.54TNEE95 pKa = 4.11VVSFPLTATVSHH107 pKa = 6.81TFSNTPVPANIQPLQISVADD127 pKa = 3.86DD128 pKa = 3.54SVNYY132 pKa = 10.03EE133 pKa = 3.98LDD135 pKa = 3.46EE136 pKa = 5.04SGTLCPTLDD145 pKa = 3.53SSVHH149 pKa = 4.13VQRR152 pKa = 11.84ATSLASALKK161 pKa = 10.46VKK163 pKa = 9.49LTGEE167 pKa = 4.13IMHH170 pKa = 6.82SDD172 pKa = 3.07SVRR175 pKa = 11.84PVQTPQLIAYY185 pKa = 8.11LFGVLLGVKK194 pKa = 10.02DD195 pKa = 3.44RR196 pKa = 11.84VNIHH200 pKa = 6.66RR201 pKa = 11.84NQPTNLWRR209 pKa = 11.84SLCSPGRR216 pKa = 11.84AAQAKK221 pKa = 9.32PFFDD225 pKa = 3.64EE226 pKa = 4.63FPNNKK231 pKa = 8.84FRR233 pKa = 11.84TGALLAPPLPDD244 pKa = 4.04AGFGPFPAEE253 pKa = 4.19GLNQNSKK260 pKa = 11.14LDD262 pKa = 3.85FRR264 pKa = 11.84SKK266 pKa = 11.0GYY268 pKa = 10.62VFYY271 pKa = 10.75KK272 pKa = 10.02QRR274 pKa = 11.84TYY276 pKa = 11.54NPDD279 pKa = 2.84EE280 pKa = 4.22MNRR283 pKa = 11.84AFWFLWAIYY292 pKa = 10.74NRR294 pKa = 11.84MPEE297 pKa = 4.43DD298 pKa = 3.51FQLSYY303 pKa = 10.62PLNITFCTSEE313 pKa = 4.99LPVQNPMPGADD324 pKa = 3.74AISNEE329 pKa = 4.08QCEE332 pKa = 4.37KK333 pKa = 11.24ALLLLEE339 pKa = 4.32KK340 pKa = 10.74VVLEE344 pKa = 4.41FFNNDD349 pKa = 2.57RR350 pKa = 11.84KK351 pKa = 10.16LAYY354 pKa = 9.58YY355 pKa = 10.1YY356 pKa = 10.73VFKK359 pKa = 10.89GCQFVMRR366 pKa = 11.84PCSCYY371 pKa = 10.16QEE373 pKa = 4.78GGLIRR378 pKa = 11.84KK379 pKa = 9.32ASRR382 pKa = 11.84NVALRR387 pKa = 11.84GFTGIYY393 pKa = 9.4YY394 pKa = 10.72LAGFTDD400 pKa = 4.1QYY402 pKa = 11.72ANMISCAVHH411 pKa = 6.09PGVVGALFQYY421 pKa = 10.39VDD423 pKa = 3.74TMVLQAVFSLSGPKK437 pKa = 9.76LVRR440 pKa = 11.84FAAPPEE446 pKa = 4.07YY447 pKa = 9.62QGRR450 pKa = 11.84HH451 pKa = 4.7ACPFSFVADD460 pKa = 3.73EE461 pKa = 4.67NYY463 pKa = 9.99WGIAPGIEE471 pKa = 3.95AEE473 pKa = 4.23PVGMYY478 pKa = 10.92YY479 pKa = 9.76MDD481 pKa = 5.05IIQRR485 pKa = 11.84KK486 pKa = 8.9AEE488 pKa = 4.0HH489 pKa = 7.02DD490 pKa = 3.84LFIEE494 pKa = 4.3TFMDD498 pKa = 3.63IYY500 pKa = 11.01GSTASIICANIEE512 pKa = 4.06TSLFTSGTNVLNHH525 pKa = 6.67RR526 pKa = 11.84MQKK529 pKa = 10.59DD530 pKa = 3.92FARR533 pKa = 11.84DD534 pKa = 3.49TPKK537 pKa = 10.66PGTLRR542 pKa = 11.84HH543 pKa = 4.59QHH545 pKa = 6.22AIINRR550 pKa = 11.84FHH552 pKa = 6.51EE553 pKa = 4.14PEE555 pKa = 3.61YY556 pKa = 10.65AYY558 pKa = 10.98RR559 pKa = 11.84LGILANGVIPLSGSFEE575 pKa = 3.98VDD577 pKa = 2.52ILKK580 pKa = 10.3EE581 pKa = 4.05AEE583 pKa = 4.13LLVTGEE589 pKa = 5.29DD590 pKa = 3.36IRR592 pKa = 11.84NLPGLRR598 pKa = 11.84CLCTRR603 pKa = 11.84GLDD606 pKa = 3.89AILGLRR612 pKa = 11.84PVQQKK617 pKa = 9.66RR618 pKa = 11.84KK619 pKa = 8.44KK620 pKa = 7.93MCYY623 pKa = 9.74FRR625 pKa = 11.84TLDD628 pKa = 3.76GNFHH632 pKa = 6.62EE633 pKa = 4.59VTIRR637 pKa = 11.84SEE639 pKa = 4.14TRR641 pKa = 11.84DD642 pKa = 3.48LQVWRR647 pKa = 11.84DD648 pKa = 3.47HH649 pKa = 6.8GYY651 pKa = 10.0LARR654 pKa = 11.84PYY656 pKa = 9.41ACHH659 pKa = 6.93IIDD662 pKa = 3.63SEE664 pKa = 4.63GIEE667 pKa = 5.36FYY669 pKa = 10.94DD670 pKa = 4.99KK671 pKa = 11.71SNGLYY676 pKa = 10.01KK677 pKa = 10.83GRR679 pKa = 11.84VNVLVSGFAIPGRR692 pKa = 11.84AYY694 pKa = 10.04QGPRR698 pKa = 11.84LQVATMAAQII708 pKa = 3.98
MM1 pKa = 7.64SAPEE5 pKa = 3.95PLNTEE10 pKa = 3.76VRR12 pKa = 11.84SPNGVSEE19 pKa = 5.13AIEE22 pKa = 4.22TQNMAVTQSSVSNEE36 pKa = 3.81IKK38 pKa = 10.69NDD40 pKa = 3.7TQSDD44 pKa = 4.17LQTLKK49 pKa = 10.89KK50 pKa = 10.1QLQPLYY56 pKa = 10.49RR57 pKa = 11.84STDD60 pKa = 3.42FNTLYY65 pKa = 11.07NFFYY69 pKa = 10.84GLDD72 pKa = 3.68VPASTDD78 pKa = 3.36RR79 pKa = 11.84VGHH82 pKa = 6.72AIQRR86 pKa = 11.84NTSVNDD92 pKa = 3.54TNEE95 pKa = 4.11VVSFPLTATVSHH107 pKa = 6.81TFSNTPVPANIQPLQISVADD127 pKa = 3.86DD128 pKa = 3.54SVNYY132 pKa = 10.03EE133 pKa = 3.98LDD135 pKa = 3.46EE136 pKa = 5.04SGTLCPTLDD145 pKa = 3.53SSVHH149 pKa = 4.13VQRR152 pKa = 11.84ATSLASALKK161 pKa = 10.46VKK163 pKa = 9.49LTGEE167 pKa = 4.13IMHH170 pKa = 6.82SDD172 pKa = 3.07SVRR175 pKa = 11.84PVQTPQLIAYY185 pKa = 8.11LFGVLLGVKK194 pKa = 10.02DD195 pKa = 3.44RR196 pKa = 11.84VNIHH200 pKa = 6.66RR201 pKa = 11.84NQPTNLWRR209 pKa = 11.84SLCSPGRR216 pKa = 11.84AAQAKK221 pKa = 9.32PFFDD225 pKa = 3.64EE226 pKa = 4.63FPNNKK231 pKa = 8.84FRR233 pKa = 11.84TGALLAPPLPDD244 pKa = 4.04AGFGPFPAEE253 pKa = 4.19GLNQNSKK260 pKa = 11.14LDD262 pKa = 3.85FRR264 pKa = 11.84SKK266 pKa = 11.0GYY268 pKa = 10.62VFYY271 pKa = 10.75KK272 pKa = 10.02QRR274 pKa = 11.84TYY276 pKa = 11.54NPDD279 pKa = 2.84EE280 pKa = 4.22MNRR283 pKa = 11.84AFWFLWAIYY292 pKa = 10.74NRR294 pKa = 11.84MPEE297 pKa = 4.43DD298 pKa = 3.51FQLSYY303 pKa = 10.62PLNITFCTSEE313 pKa = 4.99LPVQNPMPGADD324 pKa = 3.74AISNEE329 pKa = 4.08QCEE332 pKa = 4.37KK333 pKa = 11.24ALLLLEE339 pKa = 4.32KK340 pKa = 10.74VVLEE344 pKa = 4.41FFNNDD349 pKa = 2.57RR350 pKa = 11.84KK351 pKa = 10.16LAYY354 pKa = 9.58YY355 pKa = 10.1YY356 pKa = 10.73VFKK359 pKa = 10.89GCQFVMRR366 pKa = 11.84PCSCYY371 pKa = 10.16QEE373 pKa = 4.78GGLIRR378 pKa = 11.84KK379 pKa = 9.32ASRR382 pKa = 11.84NVALRR387 pKa = 11.84GFTGIYY393 pKa = 9.4YY394 pKa = 10.72LAGFTDD400 pKa = 4.1QYY402 pKa = 11.72ANMISCAVHH411 pKa = 6.09PGVVGALFQYY421 pKa = 10.39VDD423 pKa = 3.74TMVLQAVFSLSGPKK437 pKa = 9.76LVRR440 pKa = 11.84FAAPPEE446 pKa = 4.07YY447 pKa = 9.62QGRR450 pKa = 11.84HH451 pKa = 4.7ACPFSFVADD460 pKa = 3.73EE461 pKa = 4.67NYY463 pKa = 9.99WGIAPGIEE471 pKa = 3.95AEE473 pKa = 4.23PVGMYY478 pKa = 10.92YY479 pKa = 9.76MDD481 pKa = 5.05IIQRR485 pKa = 11.84KK486 pKa = 8.9AEE488 pKa = 4.0HH489 pKa = 7.02DD490 pKa = 3.84LFIEE494 pKa = 4.3TFMDD498 pKa = 3.63IYY500 pKa = 11.01GSTASIICANIEE512 pKa = 4.06TSLFTSGTNVLNHH525 pKa = 6.67RR526 pKa = 11.84MQKK529 pKa = 10.59DD530 pKa = 3.92FARR533 pKa = 11.84DD534 pKa = 3.49TPKK537 pKa = 10.66PGTLRR542 pKa = 11.84HH543 pKa = 4.59QHH545 pKa = 6.22AIINRR550 pKa = 11.84FHH552 pKa = 6.51EE553 pKa = 4.14PEE555 pKa = 3.61YY556 pKa = 10.65AYY558 pKa = 10.98RR559 pKa = 11.84LGILANGVIPLSGSFEE575 pKa = 3.98VDD577 pKa = 2.52ILKK580 pKa = 10.3EE581 pKa = 4.05AEE583 pKa = 4.13LLVTGEE589 pKa = 5.29DD590 pKa = 3.36IRR592 pKa = 11.84NLPGLRR598 pKa = 11.84CLCTRR603 pKa = 11.84GLDD606 pKa = 3.89AILGLRR612 pKa = 11.84PVQQKK617 pKa = 9.66RR618 pKa = 11.84KK619 pKa = 8.44KK620 pKa = 7.93MCYY623 pKa = 9.74FRR625 pKa = 11.84TLDD628 pKa = 3.76GNFHH632 pKa = 6.62EE633 pKa = 4.59VTIRR637 pKa = 11.84SEE639 pKa = 4.14TRR641 pKa = 11.84DD642 pKa = 3.48LQVWRR647 pKa = 11.84DD648 pKa = 3.47HH649 pKa = 6.8GYY651 pKa = 10.0LARR654 pKa = 11.84PYY656 pKa = 9.41ACHH659 pKa = 6.93IIDD662 pKa = 3.63SEE664 pKa = 4.63GIEE667 pKa = 5.36FYY669 pKa = 10.94DD670 pKa = 4.99KK671 pKa = 11.71SNGLYY676 pKa = 10.01KK677 pKa = 10.83GRR679 pKa = 11.84VNVLVSGFAIPGRR692 pKa = 11.84AYY694 pKa = 10.04QGPRR698 pKa = 11.84LQVATMAAQII708 pKa = 3.98
Molecular weight: 79.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1389 |
681 |
708 |
694.5 |
78.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.983 ± 0.448 | 2.448 ± 0.328 |
5.4 ± 0.318 | 4.752 ± 0.232 |
4.68 ± 0.577 | 5.256 ± 0.767 |
2.736 ± 0.43 | 5.976 ± 0.621 |
4.32 ± 0.451 | 9.431 ± 0.174 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.232 ± 0.079 | 5.4 ± 0.174 |
6.695 ± 0.335 | 3.96 ± 0.587 |
5.112 ± 0.473 | 7.127 ± 0.537 |
5.256 ± 0.275 | 6.695 ± 0.059 |
1.008 ± 0.21 | 4.464 ± 0.059 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
