
Plasmodium falciparum (isolate Palo Alto / Uganda)
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Apicomplexa; Aconoidasida; Haemosporida; Plasmodiidae; Plasmodium; Plasmodium (Laverania); Plasmodium falciparum
Average proteome isoelectric point is 7.27
Get precalculated fractions of proteins
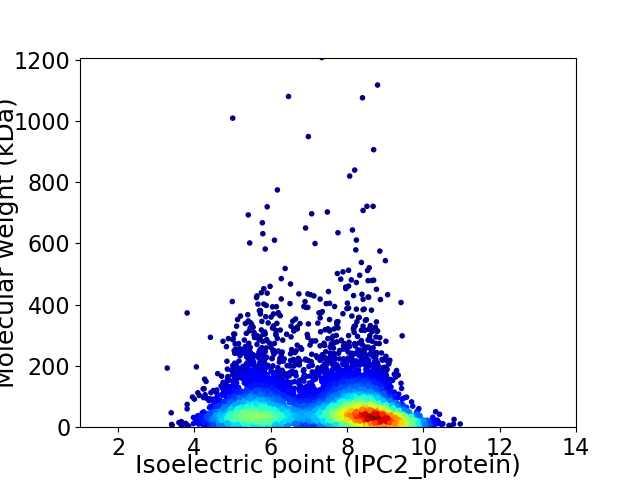
Virtual 2D-PAGE plot for 6037 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
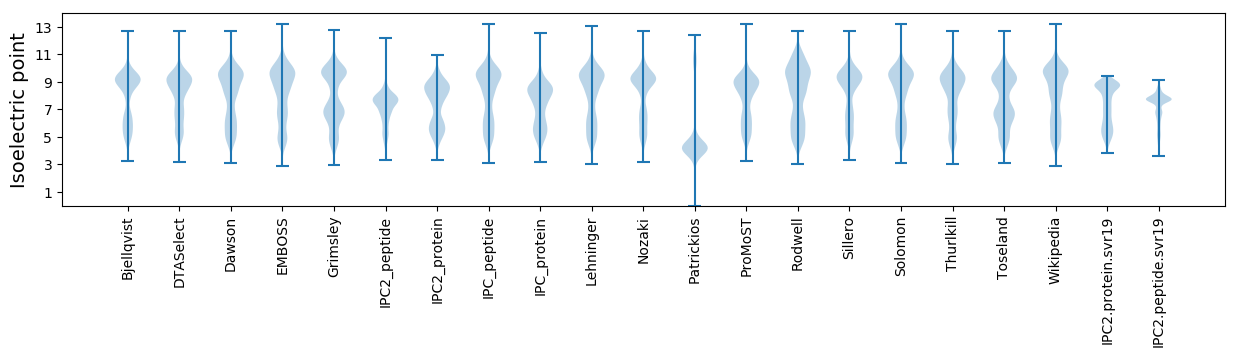
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W4J5V7|W4J5V7_PLAFP Uncharacterized protein (Fragment) OS=Plasmodium falciparum (isolate Palo Alto / Uganda) OX=57270 GN=PFUGPA_00996 PE=4 SV=1
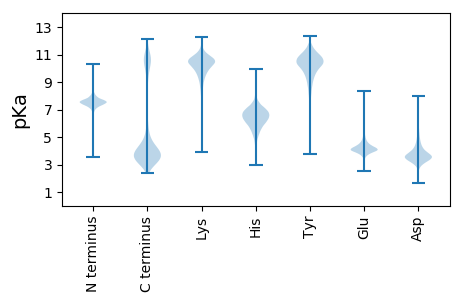
MM1 pKa = 8.65LEE3 pKa = 3.94EE4 pKa = 4.27RR5 pKa = 11.84NEE7 pKa = 3.97NNEE10 pKa = 3.93NNEE13 pKa = 4.01DD14 pKa = 3.92NEE16 pKa = 4.49NNGYY20 pKa = 8.4NQNNEE25 pKa = 3.51YY26 pKa = 10.56DD27 pKa = 3.53QNNEE31 pKa = 3.35YY32 pKa = 10.64DD33 pKa = 3.51QNNEE37 pKa = 3.35YY38 pKa = 10.64DD39 pKa = 3.51QNNEE43 pKa = 3.35YY44 pKa = 10.64DD45 pKa = 3.51QNNEE49 pKa = 3.35YY50 pKa = 10.64DD51 pKa = 3.51QNNEE55 pKa = 3.35YY56 pKa = 10.64DD57 pKa = 3.51QNNEE61 pKa = 3.37YY62 pKa = 10.72DD63 pKa = 3.43QNNDD67 pKa = 3.01YY68 pKa = 10.88NQNNDD73 pKa = 3.17YY74 pKa = 10.78NQNNDD79 pKa = 3.17YY80 pKa = 10.78NQNNDD85 pKa = 3.17YY86 pKa = 10.78NQNNDD91 pKa = 3.17YY92 pKa = 10.78NQNNDD97 pKa = 3.06YY98 pKa = 10.96NLNSGYY104 pKa = 10.55NLNSGYY110 pKa = 10.59NLNSGYY116 pKa = 10.36NLNSGYY122 pKa = 10.45SLNSGYY128 pKa = 10.98SLNNVYY134 pKa = 10.32NQYY137 pKa = 11.36SNNIEE142 pKa = 3.87NSGNILNNGFFGSSTNNNNRR162 pKa = 11.84SILGGVRR169 pKa = 11.84EE170 pKa = 4.58PIQLEE175 pKa = 4.0SRR177 pKa = 11.84GFFINGRR184 pKa = 11.84SNNVNRR190 pKa = 11.84ILVNVDD196 pKa = 3.25HH197 pKa = 7.1NNVPINSLNGTNHH210 pKa = 6.96ILGIPNNINMDD221 pKa = 4.0SLIQTILLASSLIINTSARR240 pKa = 11.84AVATSNRR247 pKa = 11.84NTRR250 pKa = 11.84NSNALTSDD258 pKa = 3.54GEE260 pKa = 4.28EE261 pKa = 4.05TMEE264 pKa = 4.89EE265 pKa = 4.03IVDD268 pKa = 4.02DD269 pKa = 4.22GMEE272 pKa = 4.32EE273 pKa = 3.7IDD275 pKa = 4.67LNEE278 pKa = 4.22NSDD281 pKa = 4.44FIHH284 pKa = 7.4DD285 pKa = 4.13GNPLGEE291 pKa = 4.28YY292 pKa = 10.96SNMEE296 pKa = 4.0YY297 pKa = 10.9EE298 pKa = 4.48LLNEE302 pKa = 4.48VSASTDD308 pKa = 3.38FSDD311 pKa = 3.5YY312 pKa = 11.01HH313 pKa = 7.75SVVDD317 pKa = 3.89EE318 pKa = 4.68DD319 pKa = 5.63LINFNDD325 pKa = 3.99DD326 pKa = 3.11VTLEE330 pKa = 4.2TQSMIAHH337 pKa = 6.92GGSLSEE343 pKa = 4.27IEE345 pKa = 4.17EE346 pKa = 4.39TGDD349 pKa = 3.66LSSDD353 pKa = 3.24VDD355 pKa = 3.74RR356 pKa = 11.84LLSSIEE362 pKa = 3.84TTPRR366 pKa = 11.84DD367 pKa = 3.48DD368 pKa = 3.26VVYY371 pKa = 10.56IRR373 pKa = 11.84GDD375 pKa = 3.33IILGEE380 pKa = 4.14DD381 pKa = 3.84EE382 pKa = 5.19IIIEE386 pKa = 4.18NGEE389 pKa = 4.12IILNNTDD396 pKa = 3.64EE397 pKa = 4.7LFEE400 pKa = 5.13DD401 pKa = 4.03VPEE404 pKa = 4.08MLEE407 pKa = 4.01NANLLEE413 pKa = 4.49DD414 pKa = 3.8RR415 pKa = 11.84EE416 pKa = 4.66HH417 pKa = 7.44VFEE420 pKa = 4.42YY421 pKa = 10.54NISVANEE428 pKa = 3.48NTTNQNSYY436 pKa = 10.71NFNAPQDD443 pKa = 4.02NTLTNYY449 pKa = 10.32NISRR453 pKa = 11.84FLQRR457 pKa = 11.84YY458 pKa = 6.69GTTTPNTQHH467 pKa = 6.75NISIHH472 pKa = 6.53RR473 pKa = 11.84DD474 pKa = 3.18GHH476 pKa = 5.2SLRR479 pKa = 11.84SDD481 pKa = 3.19TNIYY485 pKa = 10.86DD486 pKa = 4.61RR487 pKa = 11.84INGPSIRR494 pKa = 11.84EE495 pKa = 3.67NSLYY499 pKa = 10.28PGSFEE504 pKa = 3.77MAQYY508 pKa = 11.03ADD510 pKa = 4.72RR511 pKa = 11.84YY512 pKa = 10.15QMGHH516 pKa = 6.64TIAEE520 pKa = 4.43GFLYY524 pKa = 10.73
MM1 pKa = 8.65LEE3 pKa = 3.94EE4 pKa = 4.27RR5 pKa = 11.84NEE7 pKa = 3.97NNEE10 pKa = 3.93NNEE13 pKa = 4.01DD14 pKa = 3.92NEE16 pKa = 4.49NNGYY20 pKa = 8.4NQNNEE25 pKa = 3.51YY26 pKa = 10.56DD27 pKa = 3.53QNNEE31 pKa = 3.35YY32 pKa = 10.64DD33 pKa = 3.51QNNEE37 pKa = 3.35YY38 pKa = 10.64DD39 pKa = 3.51QNNEE43 pKa = 3.35YY44 pKa = 10.64DD45 pKa = 3.51QNNEE49 pKa = 3.35YY50 pKa = 10.64DD51 pKa = 3.51QNNEE55 pKa = 3.35YY56 pKa = 10.64DD57 pKa = 3.51QNNEE61 pKa = 3.37YY62 pKa = 10.72DD63 pKa = 3.43QNNDD67 pKa = 3.01YY68 pKa = 10.88NQNNDD73 pKa = 3.17YY74 pKa = 10.78NQNNDD79 pKa = 3.17YY80 pKa = 10.78NQNNDD85 pKa = 3.17YY86 pKa = 10.78NQNNDD91 pKa = 3.17YY92 pKa = 10.78NQNNDD97 pKa = 3.06YY98 pKa = 10.96NLNSGYY104 pKa = 10.55NLNSGYY110 pKa = 10.59NLNSGYY116 pKa = 10.36NLNSGYY122 pKa = 10.45SLNSGYY128 pKa = 10.98SLNNVYY134 pKa = 10.32NQYY137 pKa = 11.36SNNIEE142 pKa = 3.87NSGNILNNGFFGSSTNNNNRR162 pKa = 11.84SILGGVRR169 pKa = 11.84EE170 pKa = 4.58PIQLEE175 pKa = 4.0SRR177 pKa = 11.84GFFINGRR184 pKa = 11.84SNNVNRR190 pKa = 11.84ILVNVDD196 pKa = 3.25HH197 pKa = 7.1NNVPINSLNGTNHH210 pKa = 6.96ILGIPNNINMDD221 pKa = 4.0SLIQTILLASSLIINTSARR240 pKa = 11.84AVATSNRR247 pKa = 11.84NTRR250 pKa = 11.84NSNALTSDD258 pKa = 3.54GEE260 pKa = 4.28EE261 pKa = 4.05TMEE264 pKa = 4.89EE265 pKa = 4.03IVDD268 pKa = 4.02DD269 pKa = 4.22GMEE272 pKa = 4.32EE273 pKa = 3.7IDD275 pKa = 4.67LNEE278 pKa = 4.22NSDD281 pKa = 4.44FIHH284 pKa = 7.4DD285 pKa = 4.13GNPLGEE291 pKa = 4.28YY292 pKa = 10.96SNMEE296 pKa = 4.0YY297 pKa = 10.9EE298 pKa = 4.48LLNEE302 pKa = 4.48VSASTDD308 pKa = 3.38FSDD311 pKa = 3.5YY312 pKa = 11.01HH313 pKa = 7.75SVVDD317 pKa = 3.89EE318 pKa = 4.68DD319 pKa = 5.63LINFNDD325 pKa = 3.99DD326 pKa = 3.11VTLEE330 pKa = 4.2TQSMIAHH337 pKa = 6.92GGSLSEE343 pKa = 4.27IEE345 pKa = 4.17EE346 pKa = 4.39TGDD349 pKa = 3.66LSSDD353 pKa = 3.24VDD355 pKa = 3.74RR356 pKa = 11.84LLSSIEE362 pKa = 3.84TTPRR366 pKa = 11.84DD367 pKa = 3.48DD368 pKa = 3.26VVYY371 pKa = 10.56IRR373 pKa = 11.84GDD375 pKa = 3.33IILGEE380 pKa = 4.14DD381 pKa = 3.84EE382 pKa = 5.19IIIEE386 pKa = 4.18NGEE389 pKa = 4.12IILNNTDD396 pKa = 3.64EE397 pKa = 4.7LFEE400 pKa = 5.13DD401 pKa = 4.03VPEE404 pKa = 4.08MLEE407 pKa = 4.01NANLLEE413 pKa = 4.49DD414 pKa = 3.8RR415 pKa = 11.84EE416 pKa = 4.66HH417 pKa = 7.44VFEE420 pKa = 4.42YY421 pKa = 10.54NISVANEE428 pKa = 3.48NTTNQNSYY436 pKa = 10.71NFNAPQDD443 pKa = 4.02NTLTNYY449 pKa = 10.32NISRR453 pKa = 11.84FLQRR457 pKa = 11.84YY458 pKa = 6.69GTTTPNTQHH467 pKa = 6.75NISIHH472 pKa = 6.53RR473 pKa = 11.84DD474 pKa = 3.18GHH476 pKa = 5.2SLRR479 pKa = 11.84SDD481 pKa = 3.19TNIYY485 pKa = 10.86DD486 pKa = 4.61RR487 pKa = 11.84INGPSIRR494 pKa = 11.84EE495 pKa = 3.67NSLYY499 pKa = 10.28PGSFEE504 pKa = 3.77MAQYY508 pKa = 11.03ADD510 pKa = 4.72RR511 pKa = 11.84YY512 pKa = 10.15QMGHH516 pKa = 6.64TIAEE520 pKa = 4.43GFLYY524 pKa = 10.73
Molecular weight: 59.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W4J717|W4J717_PLAFP GpcrRhopsn4 domain-containing protein OS=Plasmodium falciparum (isolate Palo Alto / Uganda) OX=57270 GN=PFUGPA_00578 PE=4 SV=1
MM1 pKa = 7.67LGPRR5 pKa = 11.84LLQSRR10 pKa = 11.84PGLVKK15 pKa = 10.22VRR17 pKa = 11.84RR18 pKa = 11.84TLVMLGPRR26 pKa = 11.84LLQSRR31 pKa = 11.84PKK33 pKa = 10.67LVTQRR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 10.41LVMLGPIFVTLRR52 pKa = 11.84PKK54 pKa = 10.33LVNLGPIFVKK64 pKa = 10.58LRR66 pKa = 11.84PQLVNVRR73 pKa = 11.84PKK75 pKa = 10.36LVKK78 pKa = 10.54FGPMLVNVRR87 pKa = 11.84PMLVEE92 pKa = 3.87VV93 pKa = 4.11
MM1 pKa = 7.67LGPRR5 pKa = 11.84LLQSRR10 pKa = 11.84PGLVKK15 pKa = 10.22VRR17 pKa = 11.84RR18 pKa = 11.84TLVMLGPRR26 pKa = 11.84LLQSRR31 pKa = 11.84PKK33 pKa = 10.67LVTQRR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 10.41LVMLGPIFVTLRR52 pKa = 11.84PKK54 pKa = 10.33LVNLGPIFVKK64 pKa = 10.58LRR66 pKa = 11.84PQLVNVRR73 pKa = 11.84PKK75 pKa = 10.36LVKK78 pKa = 10.54FGPMLVNVRR87 pKa = 11.84PMLVEE92 pKa = 3.87VV93 pKa = 4.11
Molecular weight: 10.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3952059 |
11 |
10294 |
654.6 |
77.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
1.939 ± 0.026 | 1.787 ± 0.018 |
6.358 ± 0.029 | 6.967 ± 0.048 |
4.436 ± 0.029 | 2.793 ± 0.027 |
2.429 ± 0.014 | 9.352 ± 0.041 |
11.744 ± 0.049 | 7.642 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.217 ± 0.013 | 14.459 ± 0.104 |
1.958 ± 0.022 | 2.751 ± 0.018 |
2.644 ± 0.018 | 6.394 ± 0.025 |
4.094 ± 0.022 | 3.757 ± 0.021 |
0.497 ± 0.01 | 5.782 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
