
Hubei tombus-like virus 21
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.93
Get precalculated fractions of proteins
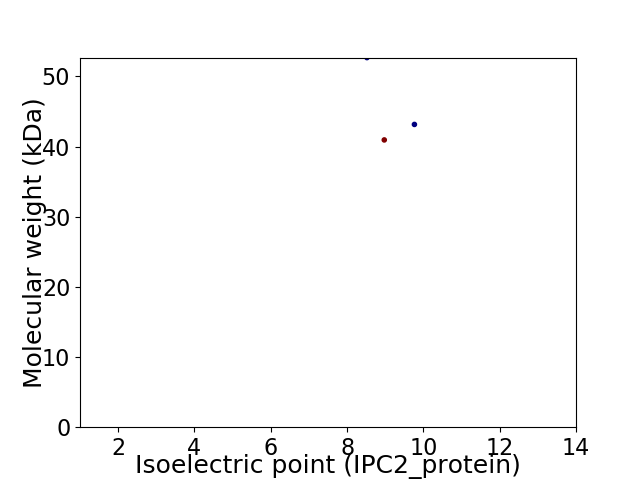
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGC9|A0A1L3KGC9_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 21 OX=1923268 PE=4 SV=1
MM1 pKa = 6.85NTVKK5 pKa = 10.74GSTFQIMTTGAKK17 pKa = 9.16PCRR20 pKa = 11.84HH21 pKa = 4.87TRR23 pKa = 11.84MLTRR27 pKa = 11.84HH28 pKa = 4.79YY29 pKa = 10.04HH30 pKa = 4.73GHH32 pKa = 6.89PLMEE36 pKa = 4.17QQHH39 pKa = 6.22IFQSCSSNEE48 pKa = 3.63YY49 pKa = 9.51RR50 pKa = 11.84GLKK53 pKa = 8.26EE54 pKa = 3.58RR55 pKa = 11.84HH56 pKa = 6.09VIEE59 pKa = 4.71QLGCSKK65 pKa = 11.15DD66 pKa = 3.78FVKK69 pKa = 10.15WAHH72 pKa = 5.41QFIAKK77 pKa = 9.89LDD79 pKa = 3.89LPSIEE84 pKa = 4.32PMSDD88 pKa = 3.03EE89 pKa = 4.28EE90 pKa = 5.15LIASRR95 pKa = 11.84PPRR98 pKa = 11.84MKK100 pKa = 10.17KK101 pKa = 10.09RR102 pKa = 11.84YY103 pKa = 9.49ASGLKK108 pKa = 10.16VPFEE112 pKa = 4.49GKK114 pKa = 8.08IHH116 pKa = 6.5ARR118 pKa = 11.84VSCFIKK124 pKa = 10.35NEE126 pKa = 3.51KK127 pKa = 8.66MAIEE131 pKa = 4.23KK132 pKa = 9.25PKK134 pKa = 9.92PRR136 pKa = 11.84CINYY140 pKa = 9.64RR141 pKa = 11.84NSIFTAQLAKK151 pKa = 8.54WTVPIEE157 pKa = 3.95KK158 pKa = 10.33LLAHH162 pKa = 6.98WPLPDD167 pKa = 3.56NGGVPFMSKK176 pKa = 9.64GRR178 pKa = 11.84NAMDD182 pKa = 4.77LGQMLNQAYY191 pKa = 7.9TLAGKK196 pKa = 10.31KK197 pKa = 9.78YY198 pKa = 9.26IHH200 pKa = 6.63LVDD203 pKa = 4.14HH204 pKa = 6.6SAYY207 pKa = 10.26DD208 pKa = 3.41GSINVEE214 pKa = 4.16HH215 pKa = 7.21IKK217 pKa = 10.88LEE219 pKa = 4.24RR220 pKa = 11.84KK221 pKa = 8.81WYY223 pKa = 10.79QKK225 pKa = 9.94MSQNDD230 pKa = 3.19SSLYY234 pKa = 10.84DD235 pKa = 4.78LITLQLEE242 pKa = 4.23NKK244 pKa = 8.61IVSRR248 pKa = 11.84NGVKK252 pKa = 10.3AKK254 pKa = 10.51CKK256 pKa = 8.78GVRR259 pKa = 11.84MSGDD263 pKa = 3.45ANTSLGNSVINYY275 pKa = 9.47IMLRR279 pKa = 11.84YY280 pKa = 9.42QYY282 pKa = 9.67PDD284 pKa = 3.15SIIIVNGDD292 pKa = 3.4DD293 pKa = 3.84SVIFSDD299 pKa = 4.54LVEE302 pKa = 4.75PAHH305 pKa = 6.12SWEE308 pKa = 4.17EE309 pKa = 3.73VGVNSKK315 pKa = 10.63VSVVQEE321 pKa = 4.14FTDD324 pKa = 4.83LEE326 pKa = 4.62YY327 pKa = 10.92CQSRR331 pKa = 11.84PVHH334 pKa = 5.47TEE336 pKa = 3.38KK337 pKa = 10.59GWVMMRR343 pKa = 11.84DD344 pKa = 4.44PIRR347 pKa = 11.84SLSRR351 pKa = 11.84MAYY354 pKa = 10.03RR355 pKa = 11.84LTHH358 pKa = 6.43GKK360 pKa = 10.38DD361 pKa = 3.13SDD363 pKa = 3.19WFYY366 pKa = 10.88TLGVGEE372 pKa = 4.3LHH374 pKa = 7.02SNPFDD379 pKa = 4.07PFMQAMAEE387 pKa = 4.14GFIKK391 pKa = 10.36RR392 pKa = 11.84GKK394 pKa = 8.68GGKK397 pKa = 8.84FRR399 pKa = 11.84SHH401 pKa = 5.56LRR403 pKa = 11.84EE404 pKa = 3.66YY405 pKa = 10.25RR406 pKa = 11.84HH407 pKa = 5.76AVGWTKK413 pKa = 10.65DD414 pKa = 3.46VEE416 pKa = 4.44RR417 pKa = 11.84ATHH420 pKa = 6.05YY421 pKa = 10.34STINWQTTFNIDD433 pKa = 3.17SAMMKK438 pKa = 10.6SMVDD442 pKa = 3.11MVLRR446 pKa = 11.84QCCYY450 pKa = 11.07ASTTSNYY457 pKa = 9.75CAA459 pKa = 4.4
MM1 pKa = 6.85NTVKK5 pKa = 10.74GSTFQIMTTGAKK17 pKa = 9.16PCRR20 pKa = 11.84HH21 pKa = 4.87TRR23 pKa = 11.84MLTRR27 pKa = 11.84HH28 pKa = 4.79YY29 pKa = 10.04HH30 pKa = 4.73GHH32 pKa = 6.89PLMEE36 pKa = 4.17QQHH39 pKa = 6.22IFQSCSSNEE48 pKa = 3.63YY49 pKa = 9.51RR50 pKa = 11.84GLKK53 pKa = 8.26EE54 pKa = 3.58RR55 pKa = 11.84HH56 pKa = 6.09VIEE59 pKa = 4.71QLGCSKK65 pKa = 11.15DD66 pKa = 3.78FVKK69 pKa = 10.15WAHH72 pKa = 5.41QFIAKK77 pKa = 9.89LDD79 pKa = 3.89LPSIEE84 pKa = 4.32PMSDD88 pKa = 3.03EE89 pKa = 4.28EE90 pKa = 5.15LIASRR95 pKa = 11.84PPRR98 pKa = 11.84MKK100 pKa = 10.17KK101 pKa = 10.09RR102 pKa = 11.84YY103 pKa = 9.49ASGLKK108 pKa = 10.16VPFEE112 pKa = 4.49GKK114 pKa = 8.08IHH116 pKa = 6.5ARR118 pKa = 11.84VSCFIKK124 pKa = 10.35NEE126 pKa = 3.51KK127 pKa = 8.66MAIEE131 pKa = 4.23KK132 pKa = 9.25PKK134 pKa = 9.92PRR136 pKa = 11.84CINYY140 pKa = 9.64RR141 pKa = 11.84NSIFTAQLAKK151 pKa = 8.54WTVPIEE157 pKa = 3.95KK158 pKa = 10.33LLAHH162 pKa = 6.98WPLPDD167 pKa = 3.56NGGVPFMSKK176 pKa = 9.64GRR178 pKa = 11.84NAMDD182 pKa = 4.77LGQMLNQAYY191 pKa = 7.9TLAGKK196 pKa = 10.31KK197 pKa = 9.78YY198 pKa = 9.26IHH200 pKa = 6.63LVDD203 pKa = 4.14HH204 pKa = 6.6SAYY207 pKa = 10.26DD208 pKa = 3.41GSINVEE214 pKa = 4.16HH215 pKa = 7.21IKK217 pKa = 10.88LEE219 pKa = 4.24RR220 pKa = 11.84KK221 pKa = 8.81WYY223 pKa = 10.79QKK225 pKa = 9.94MSQNDD230 pKa = 3.19SSLYY234 pKa = 10.84DD235 pKa = 4.78LITLQLEE242 pKa = 4.23NKK244 pKa = 8.61IVSRR248 pKa = 11.84NGVKK252 pKa = 10.3AKK254 pKa = 10.51CKK256 pKa = 8.78GVRR259 pKa = 11.84MSGDD263 pKa = 3.45ANTSLGNSVINYY275 pKa = 9.47IMLRR279 pKa = 11.84YY280 pKa = 9.42QYY282 pKa = 9.67PDD284 pKa = 3.15SIIIVNGDD292 pKa = 3.4DD293 pKa = 3.84SVIFSDD299 pKa = 4.54LVEE302 pKa = 4.75PAHH305 pKa = 6.12SWEE308 pKa = 4.17EE309 pKa = 3.73VGVNSKK315 pKa = 10.63VSVVQEE321 pKa = 4.14FTDD324 pKa = 4.83LEE326 pKa = 4.62YY327 pKa = 10.92CQSRR331 pKa = 11.84PVHH334 pKa = 5.47TEE336 pKa = 3.38KK337 pKa = 10.59GWVMMRR343 pKa = 11.84DD344 pKa = 4.44PIRR347 pKa = 11.84SLSRR351 pKa = 11.84MAYY354 pKa = 10.03RR355 pKa = 11.84LTHH358 pKa = 6.43GKK360 pKa = 10.38DD361 pKa = 3.13SDD363 pKa = 3.19WFYY366 pKa = 10.88TLGVGEE372 pKa = 4.3LHH374 pKa = 7.02SNPFDD379 pKa = 4.07PFMQAMAEE387 pKa = 4.14GFIKK391 pKa = 10.36RR392 pKa = 11.84GKK394 pKa = 8.68GGKK397 pKa = 8.84FRR399 pKa = 11.84SHH401 pKa = 5.56LRR403 pKa = 11.84EE404 pKa = 3.66YY405 pKa = 10.25RR406 pKa = 11.84HH407 pKa = 5.76AVGWTKK413 pKa = 10.65DD414 pKa = 3.46VEE416 pKa = 4.44RR417 pKa = 11.84ATHH420 pKa = 6.05YY421 pKa = 10.34STINWQTTFNIDD433 pKa = 3.17SAMMKK438 pKa = 10.6SMVDD442 pKa = 3.11MVLRR446 pKa = 11.84QCCYY450 pKa = 11.07ASTTSNYY457 pKa = 9.75CAA459 pKa = 4.4
Molecular weight: 52.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGC9|A0A1L3KGC9_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 21 OX=1923268 PE=4 SV=1
MM1 pKa = 7.39SNVGNRR7 pKa = 11.84LKK9 pKa = 10.4GTAHH13 pKa = 6.33NCASCHH19 pKa = 5.27RR20 pKa = 11.84RR21 pKa = 11.84LRR23 pKa = 11.84GQPGQRR29 pKa = 11.84FCVIVTHH36 pKa = 6.97NVDD39 pKa = 3.13QQLRR43 pKa = 11.84NLAAKK48 pKa = 9.89SWVFPNEE55 pKa = 4.0CPKK58 pKa = 10.89CGEE61 pKa = 4.18AKK63 pKa = 10.21YY64 pKa = 10.18HH65 pKa = 5.93HH66 pKa = 6.32QLANGKK72 pKa = 7.29EE73 pKa = 3.89VRR75 pKa = 11.84QCKK78 pKa = 6.85TCRR81 pKa = 11.84HH82 pKa = 5.08KK83 pKa = 10.13WGKK86 pKa = 9.32ILRR89 pKa = 11.84SSLPAKK95 pKa = 10.58SPVRR99 pKa = 11.84QPAAAPQKK107 pKa = 10.6RR108 pKa = 11.84PLCNTCMQPGMLVFGDD124 pKa = 4.44LGHH127 pKa = 7.24CLRR130 pKa = 11.84CGHH133 pKa = 5.86FTDD136 pKa = 5.36LLTLGTCSSKK146 pKa = 10.91QKK148 pKa = 10.88AKK150 pKa = 9.47AAKK153 pKa = 9.06VVKK156 pKa = 10.4HH157 pKa = 5.8NLAAPKK163 pKa = 9.1MAKK166 pKa = 8.88QAYY169 pKa = 8.19ARR171 pKa = 11.84AVEE174 pKa = 4.42EE175 pKa = 4.03RR176 pKa = 11.84LQSKK180 pKa = 8.66SFKK183 pKa = 10.52KK184 pKa = 10.12PLPTPRR190 pKa = 11.84SIFNAAKK197 pKa = 9.34PIPMPRR203 pKa = 11.84ARR205 pKa = 11.84PGPSKK210 pKa = 10.86AVVSKK215 pKa = 9.38KK216 pKa = 7.95TQVKK220 pKa = 9.48PKK222 pKa = 9.9LATIQEE228 pKa = 4.22HH229 pKa = 6.5RR230 pKa = 11.84KK231 pKa = 9.8LSPPANTKK239 pKa = 10.61KK240 pKa = 10.61LVNQQLAGTFGQPQPRR256 pKa = 11.84KK257 pKa = 10.03SRR259 pKa = 11.84DD260 pKa = 3.31SLAPRR265 pKa = 11.84ANKK268 pKa = 9.83RR269 pKa = 11.84VIRR272 pKa = 11.84AIVEE276 pKa = 4.32DD277 pKa = 4.04NNGQSDD283 pKa = 4.21VLAMRR288 pKa = 11.84KK289 pKa = 9.46AYY291 pKa = 10.17NSPQVSGNPPTKK303 pKa = 9.87EE304 pKa = 3.45DD305 pKa = 3.37FRR307 pKa = 11.84FVRR310 pKa = 11.84NLRR313 pKa = 11.84LSPHH317 pKa = 6.85LYY319 pKa = 10.15IPGEE323 pKa = 4.05GAGVWVEE330 pKa = 4.46VPALMKK336 pKa = 10.67NPRR339 pKa = 11.84LEE341 pKa = 4.22YY342 pKa = 10.9LDD344 pKa = 4.18IAWLAAMNKK353 pKa = 7.31TQEE356 pKa = 3.78QRR358 pKa = 11.84LRR360 pKa = 11.84LALILGKK367 pKa = 9.88IANQQRR373 pKa = 11.84LNKK376 pKa = 10.0AMRR379 pKa = 11.84KK380 pKa = 7.26TSPTKK385 pKa = 10.36KK386 pKa = 9.9QQ387 pKa = 2.99
MM1 pKa = 7.39SNVGNRR7 pKa = 11.84LKK9 pKa = 10.4GTAHH13 pKa = 6.33NCASCHH19 pKa = 5.27RR20 pKa = 11.84RR21 pKa = 11.84LRR23 pKa = 11.84GQPGQRR29 pKa = 11.84FCVIVTHH36 pKa = 6.97NVDD39 pKa = 3.13QQLRR43 pKa = 11.84NLAAKK48 pKa = 9.89SWVFPNEE55 pKa = 4.0CPKK58 pKa = 10.89CGEE61 pKa = 4.18AKK63 pKa = 10.21YY64 pKa = 10.18HH65 pKa = 5.93HH66 pKa = 6.32QLANGKK72 pKa = 7.29EE73 pKa = 3.89VRR75 pKa = 11.84QCKK78 pKa = 6.85TCRR81 pKa = 11.84HH82 pKa = 5.08KK83 pKa = 10.13WGKK86 pKa = 9.32ILRR89 pKa = 11.84SSLPAKK95 pKa = 10.58SPVRR99 pKa = 11.84QPAAAPQKK107 pKa = 10.6RR108 pKa = 11.84PLCNTCMQPGMLVFGDD124 pKa = 4.44LGHH127 pKa = 7.24CLRR130 pKa = 11.84CGHH133 pKa = 5.86FTDD136 pKa = 5.36LLTLGTCSSKK146 pKa = 10.91QKK148 pKa = 10.88AKK150 pKa = 9.47AAKK153 pKa = 9.06VVKK156 pKa = 10.4HH157 pKa = 5.8NLAAPKK163 pKa = 9.1MAKK166 pKa = 8.88QAYY169 pKa = 8.19ARR171 pKa = 11.84AVEE174 pKa = 4.42EE175 pKa = 4.03RR176 pKa = 11.84LQSKK180 pKa = 8.66SFKK183 pKa = 10.52KK184 pKa = 10.12PLPTPRR190 pKa = 11.84SIFNAAKK197 pKa = 9.34PIPMPRR203 pKa = 11.84ARR205 pKa = 11.84PGPSKK210 pKa = 10.86AVVSKK215 pKa = 9.38KK216 pKa = 7.95TQVKK220 pKa = 9.48PKK222 pKa = 9.9LATIQEE228 pKa = 4.22HH229 pKa = 6.5RR230 pKa = 11.84KK231 pKa = 9.8LSPPANTKK239 pKa = 10.61KK240 pKa = 10.61LVNQQLAGTFGQPQPRR256 pKa = 11.84KK257 pKa = 10.03SRR259 pKa = 11.84DD260 pKa = 3.31SLAPRR265 pKa = 11.84ANKK268 pKa = 9.83RR269 pKa = 11.84VIRR272 pKa = 11.84AIVEE276 pKa = 4.32DD277 pKa = 4.04NNGQSDD283 pKa = 4.21VLAMRR288 pKa = 11.84KK289 pKa = 9.46AYY291 pKa = 10.17NSPQVSGNPPTKK303 pKa = 9.87EE304 pKa = 3.45DD305 pKa = 3.37FRR307 pKa = 11.84FVRR310 pKa = 11.84NLRR313 pKa = 11.84LSPHH317 pKa = 6.85LYY319 pKa = 10.15IPGEE323 pKa = 4.05GAGVWVEE330 pKa = 4.46VPALMKK336 pKa = 10.67NPRR339 pKa = 11.84LEE341 pKa = 4.22YY342 pKa = 10.9LDD344 pKa = 4.18IAWLAAMNKK353 pKa = 7.31TQEE356 pKa = 3.78QRR358 pKa = 11.84LRR360 pKa = 11.84LALILGKK367 pKa = 9.88IANQQRR373 pKa = 11.84LNKK376 pKa = 10.0AMRR379 pKa = 11.84KK380 pKa = 7.26TSPTKK385 pKa = 10.36KK386 pKa = 9.9QQ387 pKa = 2.99
Molecular weight: 43.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1218 |
372 |
459 |
406.0 |
45.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.553 ± 1.15 | 1.888 ± 0.632 |
3.53 ± 0.678 | 4.105 ± 0.553 |
3.12 ± 0.311 | 6.076 ± 0.284 |
3.366 ± 0.367 | 4.68 ± 0.761 |
7.225 ± 1.439 | 7.553 ± 0.592 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.12 ± 0.795 | 4.926 ± 0.297 |
6.24 ± 1.054 | 4.68 ± 0.696 |
6.568 ± 0.664 | 7.553 ± 0.833 |
5.993 ± 1.277 | 7.143 ± 0.947 |
1.724 ± 0.271 | 2.956 ± 0.707 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
