
Cryphonectria parasitica mitovirus 1 (strain American chestnut tree/New Jersey/NB631) (CMV1) (Cryphonectria mitovirus 1)
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; Cryphonectria mitovirus 1
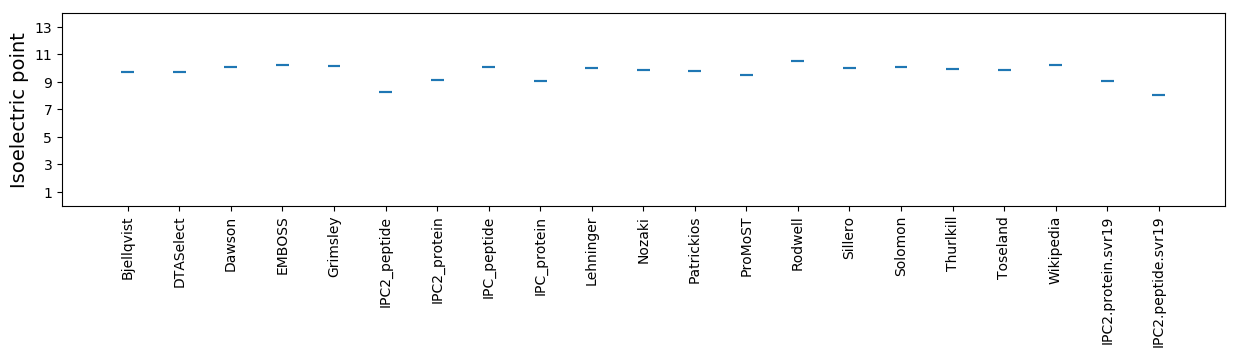
Average proteome isoelectric point is 9.08
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q33339|RDRP_CPMV1 RNA-directed RNA polymerase OS=Cryphonectria parasitica mitovirus 1 (strain American chestnut tree/New Jersey/NB631) OX=186769 PE=3 SV=1
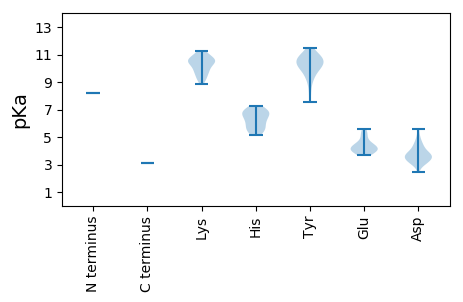
MM1 pKa = 8.21AMIIHH6 pKa = 6.88DD7 pKa = 4.35PVYY10 pKa = 10.47KK11 pKa = 10.48LWYY14 pKa = 9.18AXRR17 pKa = 11.84AKK19 pKa = 10.89SNLPGLAPHH28 pKa = 6.23KK29 pKa = 8.95TEE31 pKa = 5.34SITTVLSLTNVKK43 pKa = 10.1NKK45 pKa = 9.81QATNSILLNKK55 pKa = 8.87ITDD58 pKa = 3.71LVLGLKK64 pKa = 10.32AFSIKK69 pKa = 10.54SKK71 pKa = 10.12GVRR74 pKa = 11.84TTLRR78 pKa = 11.84VNKK81 pKa = 9.91HH82 pKa = 5.47WITPKK87 pKa = 9.94EE88 pKa = 3.85FPRR91 pKa = 11.84FVKK94 pKa = 10.51LVVWCTRR101 pKa = 11.84TQEE104 pKa = 4.43HH105 pKa = 7.23EE106 pKa = 4.19DD107 pKa = 3.41SFMKK111 pKa = 10.53IIGKK115 pKa = 9.34CDD117 pKa = 4.66HH118 pKa = 6.88IWQTAGPNFLFKK130 pKa = 10.73YY131 pKa = 9.13LKK133 pKa = 9.98EE134 pKa = 4.08VMRR137 pKa = 11.84LSVRR141 pKa = 11.84RR142 pKa = 11.84IANIEE147 pKa = 4.0LEE149 pKa = 4.12PSKK152 pKa = 11.06KK153 pKa = 10.23IFVKK157 pKa = 10.39LNKK160 pKa = 9.32FRR162 pKa = 11.84FPNIIPLPICDD173 pKa = 4.86QIIRR177 pKa = 11.84DD178 pKa = 3.83QNDD181 pKa = 2.97QVLWASKK188 pKa = 10.63RR189 pKa = 11.84LIICLLTILSVHH201 pKa = 6.19RR202 pKa = 11.84VLPTKK207 pKa = 10.18VVPDD211 pKa = 3.47YY212 pKa = 10.86STIVDD217 pKa = 4.14PFTGVSKK224 pKa = 9.81TIDD227 pKa = 3.09QKK229 pKa = 11.21LLRR232 pKa = 11.84KK233 pKa = 9.82AIHH236 pKa = 6.42LLNIKK241 pKa = 9.6RR242 pKa = 11.84VKK244 pKa = 9.5QLKK247 pKa = 10.31LKK249 pKa = 9.18ITGSMKK255 pKa = 10.45AGPNGKK261 pKa = 9.21ISLLTSSVDD270 pKa = 3.1ALSFITQPTKK280 pKa = 10.46IFTYY284 pKa = 10.08LDD286 pKa = 3.36FSVRR290 pKa = 11.84VYY292 pKa = 11.03KK293 pKa = 10.76FRR295 pKa = 11.84GLLLWMWMMCILLITLPYY313 pKa = 10.25AIVSFMLGALIPIMGKK329 pKa = 10.33LSVVYY334 pKa = 10.13DD335 pKa = 3.57QAGKK339 pKa = 10.67ARR341 pKa = 11.84IVAITNSWIQTAFYY355 pKa = 10.5SLHH358 pKa = 5.63LHH360 pKa = 5.62VFKK363 pKa = 10.89LLKK366 pKa = 10.92NIDD369 pKa = 3.34QDD371 pKa = 3.76GTFDD375 pKa = 3.45QEE377 pKa = 4.53RR378 pKa = 11.84PFKK381 pKa = 10.96LLIKK385 pKa = 9.25WLNEE389 pKa = 3.66PTQKK393 pKa = 10.58FYY395 pKa = 11.47GFDD398 pKa = 3.07LTAATDD404 pKa = 3.71RR405 pKa = 11.84LPIDD409 pKa = 3.86LQVDD413 pKa = 3.68ILNIIFKK420 pKa = 10.32NSPGSSWRR428 pKa = 11.84SLLRR432 pKa = 11.84IKK434 pKa = 10.69YY435 pKa = 9.48KK436 pKa = 10.62SPQGFLTYY444 pKa = 10.76AVGQPMGAYY453 pKa = 10.13SSFAMLALTHH463 pKa = 6.12HH464 pKa = 6.86VIVQVAALNSGFTTRR479 pKa = 11.84FTDD482 pKa = 3.65YY483 pKa = 10.87CILGDD488 pKa = 5.09DD489 pKa = 4.01IVIAHH494 pKa = 6.64DD495 pKa = 4.06TVASEE500 pKa = 3.98YY501 pKa = 10.49LKK503 pKa = 11.23LMEE506 pKa = 4.54TLGLSISSGKK516 pKa = 10.24SVISSEE522 pKa = 3.82FTEE525 pKa = 4.71FAKK528 pKa = 10.72KK529 pKa = 10.55LKK531 pKa = 10.04GRR533 pKa = 11.84NNFDD537 pKa = 2.43IFYY540 pKa = 9.55RR541 pKa = 11.84SWFSIIHH548 pKa = 5.45FEE550 pKa = 4.22KK551 pKa = 10.67QILHH555 pKa = 6.65LCTVFEE561 pKa = 4.17LLRR564 pKa = 11.84RR565 pKa = 11.84GVCEE569 pKa = 5.59LYY571 pKa = 10.66DD572 pKa = 5.57LYY574 pKa = 10.56PQYY577 pKa = 10.96INKK580 pKa = 9.48LPKK583 pKa = 9.66IYY585 pKa = 10.46LRR587 pKa = 11.84YY588 pKa = 9.94NLLIDD593 pKa = 3.56WVVVAFTNQILIGDD607 pKa = 4.52RR608 pKa = 11.84PRR610 pKa = 11.84ADD612 pKa = 4.34GIRR615 pKa = 11.84LFDD618 pKa = 3.64YY619 pKa = 10.59FVGLEE624 pKa = 4.2VIPPLLRR631 pKa = 11.84IMLHH635 pKa = 5.86TIKK638 pKa = 10.56KK639 pKa = 9.61DD640 pKa = 3.37WNGLWNSIKK649 pKa = 9.69YY650 pKa = 7.53TLNKK654 pKa = 9.72GFVVSQVRR662 pKa = 11.84VGLPDD667 pKa = 3.09WTEE670 pKa = 3.92LFLLPILPSTYY681 pKa = 9.9IIIRR685 pKa = 11.84DD686 pKa = 3.6YY687 pKa = 11.19CRR689 pKa = 11.84SFNDD693 pKa = 3.28LTKK696 pKa = 11.03LFGEE700 pKa = 4.17WWLLRR705 pKa = 11.84FEE707 pKa = 4.33SEE709 pKa = 4.45SYY711 pKa = 10.06QVSILDD717 pKa = 3.8VIDD720 pKa = 4.45RR721 pKa = 11.84LAHH724 pKa = 5.15TSIPNLDD731 pKa = 3.02IHH733 pKa = 6.99DD734 pKa = 4.12KK735 pKa = 11.14KK736 pKa = 10.84KK737 pKa = 11.27VKK739 pKa = 9.15LTLDD743 pKa = 3.43NLYY746 pKa = 10.72KK747 pKa = 10.8LSLIVNIPSGGARR760 pKa = 11.84RR761 pKa = 11.84YY762 pKa = 10.1IEE764 pKa = 3.73FLRR767 pKa = 11.84FNGLKK772 pKa = 10.34SPLIVEE778 pKa = 5.18RR779 pKa = 11.84YY780 pKa = 8.4IKK782 pKa = 10.64DD783 pKa = 3.74GIRR786 pKa = 11.84IEE788 pKa = 5.0KK789 pKa = 10.0PLTLQGLHH797 pKa = 6.77RR798 pKa = 11.84SGDD801 pKa = 3.28IQLGFKK807 pKa = 10.36ISS809 pKa = 3.13
MM1 pKa = 8.21AMIIHH6 pKa = 6.88DD7 pKa = 4.35PVYY10 pKa = 10.47KK11 pKa = 10.48LWYY14 pKa = 9.18AXRR17 pKa = 11.84AKK19 pKa = 10.89SNLPGLAPHH28 pKa = 6.23KK29 pKa = 8.95TEE31 pKa = 5.34SITTVLSLTNVKK43 pKa = 10.1NKK45 pKa = 9.81QATNSILLNKK55 pKa = 8.87ITDD58 pKa = 3.71LVLGLKK64 pKa = 10.32AFSIKK69 pKa = 10.54SKK71 pKa = 10.12GVRR74 pKa = 11.84TTLRR78 pKa = 11.84VNKK81 pKa = 9.91HH82 pKa = 5.47WITPKK87 pKa = 9.94EE88 pKa = 3.85FPRR91 pKa = 11.84FVKK94 pKa = 10.51LVVWCTRR101 pKa = 11.84TQEE104 pKa = 4.43HH105 pKa = 7.23EE106 pKa = 4.19DD107 pKa = 3.41SFMKK111 pKa = 10.53IIGKK115 pKa = 9.34CDD117 pKa = 4.66HH118 pKa = 6.88IWQTAGPNFLFKK130 pKa = 10.73YY131 pKa = 9.13LKK133 pKa = 9.98EE134 pKa = 4.08VMRR137 pKa = 11.84LSVRR141 pKa = 11.84RR142 pKa = 11.84IANIEE147 pKa = 4.0LEE149 pKa = 4.12PSKK152 pKa = 11.06KK153 pKa = 10.23IFVKK157 pKa = 10.39LNKK160 pKa = 9.32FRR162 pKa = 11.84FPNIIPLPICDD173 pKa = 4.86QIIRR177 pKa = 11.84DD178 pKa = 3.83QNDD181 pKa = 2.97QVLWASKK188 pKa = 10.63RR189 pKa = 11.84LIICLLTILSVHH201 pKa = 6.19RR202 pKa = 11.84VLPTKK207 pKa = 10.18VVPDD211 pKa = 3.47YY212 pKa = 10.86STIVDD217 pKa = 4.14PFTGVSKK224 pKa = 9.81TIDD227 pKa = 3.09QKK229 pKa = 11.21LLRR232 pKa = 11.84KK233 pKa = 9.82AIHH236 pKa = 6.42LLNIKK241 pKa = 9.6RR242 pKa = 11.84VKK244 pKa = 9.5QLKK247 pKa = 10.31LKK249 pKa = 9.18ITGSMKK255 pKa = 10.45AGPNGKK261 pKa = 9.21ISLLTSSVDD270 pKa = 3.1ALSFITQPTKK280 pKa = 10.46IFTYY284 pKa = 10.08LDD286 pKa = 3.36FSVRR290 pKa = 11.84VYY292 pKa = 11.03KK293 pKa = 10.76FRR295 pKa = 11.84GLLLWMWMMCILLITLPYY313 pKa = 10.25AIVSFMLGALIPIMGKK329 pKa = 10.33LSVVYY334 pKa = 10.13DD335 pKa = 3.57QAGKK339 pKa = 10.67ARR341 pKa = 11.84IVAITNSWIQTAFYY355 pKa = 10.5SLHH358 pKa = 5.63LHH360 pKa = 5.62VFKK363 pKa = 10.89LLKK366 pKa = 10.92NIDD369 pKa = 3.34QDD371 pKa = 3.76GTFDD375 pKa = 3.45QEE377 pKa = 4.53RR378 pKa = 11.84PFKK381 pKa = 10.96LLIKK385 pKa = 9.25WLNEE389 pKa = 3.66PTQKK393 pKa = 10.58FYY395 pKa = 11.47GFDD398 pKa = 3.07LTAATDD404 pKa = 3.71RR405 pKa = 11.84LPIDD409 pKa = 3.86LQVDD413 pKa = 3.68ILNIIFKK420 pKa = 10.32NSPGSSWRR428 pKa = 11.84SLLRR432 pKa = 11.84IKK434 pKa = 10.69YY435 pKa = 9.48KK436 pKa = 10.62SPQGFLTYY444 pKa = 10.76AVGQPMGAYY453 pKa = 10.13SSFAMLALTHH463 pKa = 6.12HH464 pKa = 6.86VIVQVAALNSGFTTRR479 pKa = 11.84FTDD482 pKa = 3.65YY483 pKa = 10.87CILGDD488 pKa = 5.09DD489 pKa = 4.01IVIAHH494 pKa = 6.64DD495 pKa = 4.06TVASEE500 pKa = 3.98YY501 pKa = 10.49LKK503 pKa = 11.23LMEE506 pKa = 4.54TLGLSISSGKK516 pKa = 10.24SVISSEE522 pKa = 3.82FTEE525 pKa = 4.71FAKK528 pKa = 10.72KK529 pKa = 10.55LKK531 pKa = 10.04GRR533 pKa = 11.84NNFDD537 pKa = 2.43IFYY540 pKa = 9.55RR541 pKa = 11.84SWFSIIHH548 pKa = 5.45FEE550 pKa = 4.22KK551 pKa = 10.67QILHH555 pKa = 6.65LCTVFEE561 pKa = 4.17LLRR564 pKa = 11.84RR565 pKa = 11.84GVCEE569 pKa = 5.59LYY571 pKa = 10.66DD572 pKa = 5.57LYY574 pKa = 10.56PQYY577 pKa = 10.96INKK580 pKa = 9.48LPKK583 pKa = 9.66IYY585 pKa = 10.46LRR587 pKa = 11.84YY588 pKa = 9.94NLLIDD593 pKa = 3.56WVVVAFTNQILIGDD607 pKa = 4.52RR608 pKa = 11.84PRR610 pKa = 11.84ADD612 pKa = 4.34GIRR615 pKa = 11.84LFDD618 pKa = 3.64YY619 pKa = 10.59FVGLEE624 pKa = 4.2VIPPLLRR631 pKa = 11.84IMLHH635 pKa = 5.86TIKK638 pKa = 10.56KK639 pKa = 9.61DD640 pKa = 3.37WNGLWNSIKK649 pKa = 9.69YY650 pKa = 7.53TLNKK654 pKa = 9.72GFVVSQVRR662 pKa = 11.84VGLPDD667 pKa = 3.09WTEE670 pKa = 3.92LFLLPILPSTYY681 pKa = 9.9IIIRR685 pKa = 11.84DD686 pKa = 3.6YY687 pKa = 11.19CRR689 pKa = 11.84SFNDD693 pKa = 3.28LTKK696 pKa = 11.03LFGEE700 pKa = 4.17WWLLRR705 pKa = 11.84FEE707 pKa = 4.33SEE709 pKa = 4.45SYY711 pKa = 10.06QVSILDD717 pKa = 3.8VIDD720 pKa = 4.45RR721 pKa = 11.84LAHH724 pKa = 5.15TSIPNLDD731 pKa = 3.02IHH733 pKa = 6.99DD734 pKa = 4.12KK735 pKa = 11.14KK736 pKa = 10.84KK737 pKa = 11.27VKK739 pKa = 9.15LTLDD743 pKa = 3.43NLYY746 pKa = 10.72KK747 pKa = 10.8LSLIVNIPSGGARR760 pKa = 11.84RR761 pKa = 11.84YY762 pKa = 10.1IEE764 pKa = 3.73FLRR767 pKa = 11.84FNGLKK772 pKa = 10.34SPLIVEE778 pKa = 5.18RR779 pKa = 11.84YY780 pKa = 8.4IKK782 pKa = 10.64DD783 pKa = 3.74GIRR786 pKa = 11.84IEE788 pKa = 5.0KK789 pKa = 10.0PLTLQGLHH797 pKa = 6.77RR798 pKa = 11.84SGDD801 pKa = 3.28IQLGFKK807 pKa = 10.36ISS809 pKa = 3.13
Molecular weight: 93.07 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q33339|RDRP_CPMV1 RNA-directed RNA polymerase OS=Cryphonectria parasitica mitovirus 1 (strain American chestnut tree/New Jersey/NB631) OX=186769 PE=3 SV=1
MM1 pKa = 8.21AMIIHH6 pKa = 6.88DD7 pKa = 4.35PVYY10 pKa = 10.47KK11 pKa = 10.48LWYY14 pKa = 9.18AXRR17 pKa = 11.84AKK19 pKa = 10.89SNLPGLAPHH28 pKa = 6.23KK29 pKa = 8.95TEE31 pKa = 5.34SITTVLSLTNVKK43 pKa = 10.1NKK45 pKa = 9.81QATNSILLNKK55 pKa = 8.87ITDD58 pKa = 3.71LVLGLKK64 pKa = 10.32AFSIKK69 pKa = 10.54SKK71 pKa = 10.12GVRR74 pKa = 11.84TTLRR78 pKa = 11.84VNKK81 pKa = 9.91HH82 pKa = 5.47WITPKK87 pKa = 9.94EE88 pKa = 3.85FPRR91 pKa = 11.84FVKK94 pKa = 10.51LVVWCTRR101 pKa = 11.84TQEE104 pKa = 4.43HH105 pKa = 7.23EE106 pKa = 4.19DD107 pKa = 3.41SFMKK111 pKa = 10.53IIGKK115 pKa = 9.34CDD117 pKa = 4.66HH118 pKa = 6.88IWQTAGPNFLFKK130 pKa = 10.73YY131 pKa = 9.13LKK133 pKa = 9.98EE134 pKa = 4.08VMRR137 pKa = 11.84LSVRR141 pKa = 11.84RR142 pKa = 11.84IANIEE147 pKa = 4.0LEE149 pKa = 4.12PSKK152 pKa = 11.06KK153 pKa = 10.23IFVKK157 pKa = 10.39LNKK160 pKa = 9.32FRR162 pKa = 11.84FPNIIPLPICDD173 pKa = 4.86QIIRR177 pKa = 11.84DD178 pKa = 3.83QNDD181 pKa = 2.97QVLWASKK188 pKa = 10.63RR189 pKa = 11.84LIICLLTILSVHH201 pKa = 6.19RR202 pKa = 11.84VLPTKK207 pKa = 10.18VVPDD211 pKa = 3.47YY212 pKa = 10.86STIVDD217 pKa = 4.14PFTGVSKK224 pKa = 9.81TIDD227 pKa = 3.09QKK229 pKa = 11.21LLRR232 pKa = 11.84KK233 pKa = 9.82AIHH236 pKa = 6.42LLNIKK241 pKa = 9.6RR242 pKa = 11.84VKK244 pKa = 9.5QLKK247 pKa = 10.31LKK249 pKa = 9.18ITGSMKK255 pKa = 10.45AGPNGKK261 pKa = 9.21ISLLTSSVDD270 pKa = 3.1ALSFITQPTKK280 pKa = 10.46IFTYY284 pKa = 10.08LDD286 pKa = 3.36FSVRR290 pKa = 11.84VYY292 pKa = 11.03KK293 pKa = 10.76FRR295 pKa = 11.84GLLLWMWMMCILLITLPYY313 pKa = 10.25AIVSFMLGALIPIMGKK329 pKa = 10.33LSVVYY334 pKa = 10.13DD335 pKa = 3.57QAGKK339 pKa = 10.67ARR341 pKa = 11.84IVAITNSWIQTAFYY355 pKa = 10.5SLHH358 pKa = 5.63LHH360 pKa = 5.62VFKK363 pKa = 10.89LLKK366 pKa = 10.92NIDD369 pKa = 3.34QDD371 pKa = 3.76GTFDD375 pKa = 3.45QEE377 pKa = 4.53RR378 pKa = 11.84PFKK381 pKa = 10.96LLIKK385 pKa = 9.25WLNEE389 pKa = 3.66PTQKK393 pKa = 10.58FYY395 pKa = 11.47GFDD398 pKa = 3.07LTAATDD404 pKa = 3.71RR405 pKa = 11.84LPIDD409 pKa = 3.86LQVDD413 pKa = 3.68ILNIIFKK420 pKa = 10.32NSPGSSWRR428 pKa = 11.84SLLRR432 pKa = 11.84IKK434 pKa = 10.69YY435 pKa = 9.48KK436 pKa = 10.62SPQGFLTYY444 pKa = 10.76AVGQPMGAYY453 pKa = 10.13SSFAMLALTHH463 pKa = 6.12HH464 pKa = 6.86VIVQVAALNSGFTTRR479 pKa = 11.84FTDD482 pKa = 3.65YY483 pKa = 10.87CILGDD488 pKa = 5.09DD489 pKa = 4.01IVIAHH494 pKa = 6.64DD495 pKa = 4.06TVASEE500 pKa = 3.98YY501 pKa = 10.49LKK503 pKa = 11.23LMEE506 pKa = 4.54TLGLSISSGKK516 pKa = 10.24SVISSEE522 pKa = 3.82FTEE525 pKa = 4.71FAKK528 pKa = 10.72KK529 pKa = 10.55LKK531 pKa = 10.04GRR533 pKa = 11.84NNFDD537 pKa = 2.43IFYY540 pKa = 9.55RR541 pKa = 11.84SWFSIIHH548 pKa = 5.45FEE550 pKa = 4.22KK551 pKa = 10.67QILHH555 pKa = 6.65LCTVFEE561 pKa = 4.17LLRR564 pKa = 11.84RR565 pKa = 11.84GVCEE569 pKa = 5.59LYY571 pKa = 10.66DD572 pKa = 5.57LYY574 pKa = 10.56PQYY577 pKa = 10.96INKK580 pKa = 9.48LPKK583 pKa = 9.66IYY585 pKa = 10.46LRR587 pKa = 11.84YY588 pKa = 9.94NLLIDD593 pKa = 3.56WVVVAFTNQILIGDD607 pKa = 4.52RR608 pKa = 11.84PRR610 pKa = 11.84ADD612 pKa = 4.34GIRR615 pKa = 11.84LFDD618 pKa = 3.64YY619 pKa = 10.59FVGLEE624 pKa = 4.2VIPPLLRR631 pKa = 11.84IMLHH635 pKa = 5.86TIKK638 pKa = 10.56KK639 pKa = 9.61DD640 pKa = 3.37WNGLWNSIKK649 pKa = 9.69YY650 pKa = 7.53TLNKK654 pKa = 9.72GFVVSQVRR662 pKa = 11.84VGLPDD667 pKa = 3.09WTEE670 pKa = 3.92LFLLPILPSTYY681 pKa = 9.9IIIRR685 pKa = 11.84DD686 pKa = 3.6YY687 pKa = 11.19CRR689 pKa = 11.84SFNDD693 pKa = 3.28LTKK696 pKa = 11.03LFGEE700 pKa = 4.17WWLLRR705 pKa = 11.84FEE707 pKa = 4.33SEE709 pKa = 4.45SYY711 pKa = 10.06QVSILDD717 pKa = 3.8VIDD720 pKa = 4.45RR721 pKa = 11.84LAHH724 pKa = 5.15TSIPNLDD731 pKa = 3.02IHH733 pKa = 6.99DD734 pKa = 4.12KK735 pKa = 11.14KK736 pKa = 10.84KK737 pKa = 11.27VKK739 pKa = 9.15LTLDD743 pKa = 3.43NLYY746 pKa = 10.72KK747 pKa = 10.8LSLIVNIPSGGARR760 pKa = 11.84RR761 pKa = 11.84YY762 pKa = 10.1IEE764 pKa = 3.73FLRR767 pKa = 11.84FNGLKK772 pKa = 10.34SPLIVEE778 pKa = 5.18RR779 pKa = 11.84YY780 pKa = 8.4IKK782 pKa = 10.64DD783 pKa = 3.74GIRR786 pKa = 11.84IEE788 pKa = 5.0KK789 pKa = 10.0PLTLQGLHH797 pKa = 6.77RR798 pKa = 11.84SGDD801 pKa = 3.28IQLGFKK807 pKa = 10.36ISS809 pKa = 3.13
MM1 pKa = 8.21AMIIHH6 pKa = 6.88DD7 pKa = 4.35PVYY10 pKa = 10.47KK11 pKa = 10.48LWYY14 pKa = 9.18AXRR17 pKa = 11.84AKK19 pKa = 10.89SNLPGLAPHH28 pKa = 6.23KK29 pKa = 8.95TEE31 pKa = 5.34SITTVLSLTNVKK43 pKa = 10.1NKK45 pKa = 9.81QATNSILLNKK55 pKa = 8.87ITDD58 pKa = 3.71LVLGLKK64 pKa = 10.32AFSIKK69 pKa = 10.54SKK71 pKa = 10.12GVRR74 pKa = 11.84TTLRR78 pKa = 11.84VNKK81 pKa = 9.91HH82 pKa = 5.47WITPKK87 pKa = 9.94EE88 pKa = 3.85FPRR91 pKa = 11.84FVKK94 pKa = 10.51LVVWCTRR101 pKa = 11.84TQEE104 pKa = 4.43HH105 pKa = 7.23EE106 pKa = 4.19DD107 pKa = 3.41SFMKK111 pKa = 10.53IIGKK115 pKa = 9.34CDD117 pKa = 4.66HH118 pKa = 6.88IWQTAGPNFLFKK130 pKa = 10.73YY131 pKa = 9.13LKK133 pKa = 9.98EE134 pKa = 4.08VMRR137 pKa = 11.84LSVRR141 pKa = 11.84RR142 pKa = 11.84IANIEE147 pKa = 4.0LEE149 pKa = 4.12PSKK152 pKa = 11.06KK153 pKa = 10.23IFVKK157 pKa = 10.39LNKK160 pKa = 9.32FRR162 pKa = 11.84FPNIIPLPICDD173 pKa = 4.86QIIRR177 pKa = 11.84DD178 pKa = 3.83QNDD181 pKa = 2.97QVLWASKK188 pKa = 10.63RR189 pKa = 11.84LIICLLTILSVHH201 pKa = 6.19RR202 pKa = 11.84VLPTKK207 pKa = 10.18VVPDD211 pKa = 3.47YY212 pKa = 10.86STIVDD217 pKa = 4.14PFTGVSKK224 pKa = 9.81TIDD227 pKa = 3.09QKK229 pKa = 11.21LLRR232 pKa = 11.84KK233 pKa = 9.82AIHH236 pKa = 6.42LLNIKK241 pKa = 9.6RR242 pKa = 11.84VKK244 pKa = 9.5QLKK247 pKa = 10.31LKK249 pKa = 9.18ITGSMKK255 pKa = 10.45AGPNGKK261 pKa = 9.21ISLLTSSVDD270 pKa = 3.1ALSFITQPTKK280 pKa = 10.46IFTYY284 pKa = 10.08LDD286 pKa = 3.36FSVRR290 pKa = 11.84VYY292 pKa = 11.03KK293 pKa = 10.76FRR295 pKa = 11.84GLLLWMWMMCILLITLPYY313 pKa = 10.25AIVSFMLGALIPIMGKK329 pKa = 10.33LSVVYY334 pKa = 10.13DD335 pKa = 3.57QAGKK339 pKa = 10.67ARR341 pKa = 11.84IVAITNSWIQTAFYY355 pKa = 10.5SLHH358 pKa = 5.63LHH360 pKa = 5.62VFKK363 pKa = 10.89LLKK366 pKa = 10.92NIDD369 pKa = 3.34QDD371 pKa = 3.76GTFDD375 pKa = 3.45QEE377 pKa = 4.53RR378 pKa = 11.84PFKK381 pKa = 10.96LLIKK385 pKa = 9.25WLNEE389 pKa = 3.66PTQKK393 pKa = 10.58FYY395 pKa = 11.47GFDD398 pKa = 3.07LTAATDD404 pKa = 3.71RR405 pKa = 11.84LPIDD409 pKa = 3.86LQVDD413 pKa = 3.68ILNIIFKK420 pKa = 10.32NSPGSSWRR428 pKa = 11.84SLLRR432 pKa = 11.84IKK434 pKa = 10.69YY435 pKa = 9.48KK436 pKa = 10.62SPQGFLTYY444 pKa = 10.76AVGQPMGAYY453 pKa = 10.13SSFAMLALTHH463 pKa = 6.12HH464 pKa = 6.86VIVQVAALNSGFTTRR479 pKa = 11.84FTDD482 pKa = 3.65YY483 pKa = 10.87CILGDD488 pKa = 5.09DD489 pKa = 4.01IVIAHH494 pKa = 6.64DD495 pKa = 4.06TVASEE500 pKa = 3.98YY501 pKa = 10.49LKK503 pKa = 11.23LMEE506 pKa = 4.54TLGLSISSGKK516 pKa = 10.24SVISSEE522 pKa = 3.82FTEE525 pKa = 4.71FAKK528 pKa = 10.72KK529 pKa = 10.55LKK531 pKa = 10.04GRR533 pKa = 11.84NNFDD537 pKa = 2.43IFYY540 pKa = 9.55RR541 pKa = 11.84SWFSIIHH548 pKa = 5.45FEE550 pKa = 4.22KK551 pKa = 10.67QILHH555 pKa = 6.65LCTVFEE561 pKa = 4.17LLRR564 pKa = 11.84RR565 pKa = 11.84GVCEE569 pKa = 5.59LYY571 pKa = 10.66DD572 pKa = 5.57LYY574 pKa = 10.56PQYY577 pKa = 10.96INKK580 pKa = 9.48LPKK583 pKa = 9.66IYY585 pKa = 10.46LRR587 pKa = 11.84YY588 pKa = 9.94NLLIDD593 pKa = 3.56WVVVAFTNQILIGDD607 pKa = 4.52RR608 pKa = 11.84PRR610 pKa = 11.84ADD612 pKa = 4.34GIRR615 pKa = 11.84LFDD618 pKa = 3.64YY619 pKa = 10.59FVGLEE624 pKa = 4.2VIPPLLRR631 pKa = 11.84IMLHH635 pKa = 5.86TIKK638 pKa = 10.56KK639 pKa = 9.61DD640 pKa = 3.37WNGLWNSIKK649 pKa = 9.69YY650 pKa = 7.53TLNKK654 pKa = 9.72GFVVSQVRR662 pKa = 11.84VGLPDD667 pKa = 3.09WTEE670 pKa = 3.92LFLLPILPSTYY681 pKa = 9.9IIIRR685 pKa = 11.84DD686 pKa = 3.6YY687 pKa = 11.19CRR689 pKa = 11.84SFNDD693 pKa = 3.28LTKK696 pKa = 11.03LFGEE700 pKa = 4.17WWLLRR705 pKa = 11.84FEE707 pKa = 4.33SEE709 pKa = 4.45SYY711 pKa = 10.06QVSILDD717 pKa = 3.8VIDD720 pKa = 4.45RR721 pKa = 11.84LAHH724 pKa = 5.15TSIPNLDD731 pKa = 3.02IHH733 pKa = 6.99DD734 pKa = 4.12KK735 pKa = 11.14KK736 pKa = 10.84KK737 pKa = 11.27VKK739 pKa = 9.15LTLDD743 pKa = 3.43NLYY746 pKa = 10.72KK747 pKa = 10.8LSLIVNIPSGGARR760 pKa = 11.84RR761 pKa = 11.84YY762 pKa = 10.1IEE764 pKa = 3.73FLRR767 pKa = 11.84FNGLKK772 pKa = 10.34SPLIVEE778 pKa = 5.18RR779 pKa = 11.84YY780 pKa = 8.4IKK782 pKa = 10.64DD783 pKa = 3.74GIRR786 pKa = 11.84IEE788 pKa = 5.0KK789 pKa = 10.0PLTLQGLHH797 pKa = 6.77RR798 pKa = 11.84SGDD801 pKa = 3.28IQLGFKK807 pKa = 10.36ISS809 pKa = 3.13
Molecular weight: 93.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
809 |
809 |
809 |
809.0 |
93.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.079 ± 0.0 | 1.112 ± 0.0 |
5.068 ± 0.0 | 2.967 ± 0.0 |
5.439 ± 0.0 | 4.821 ± 0.0 |
2.225 ± 0.0 | 10.012 ± 0.0 |
8.035 ± 0.0 | 13.35 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.731 ± 0.0 | 3.956 ± 0.0 |
4.326 ± 0.0 | 3.09 ± 0.0 |
5.192 ± 0.0 | 6.551 ± 0.0 |
5.81 ± 0.0 | 6.428 ± 0.0 |
2.101 ± 0.0 | 3.585 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
