
Dragonfly larvae associated circular virus-9
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
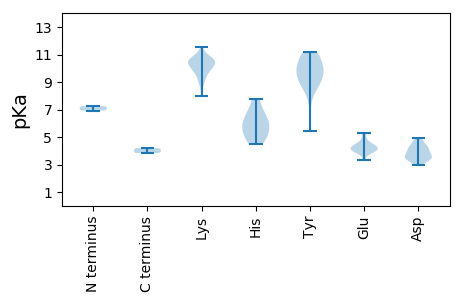
Average proteome isoelectric point is 8.14
Get precalculated fractions of proteins
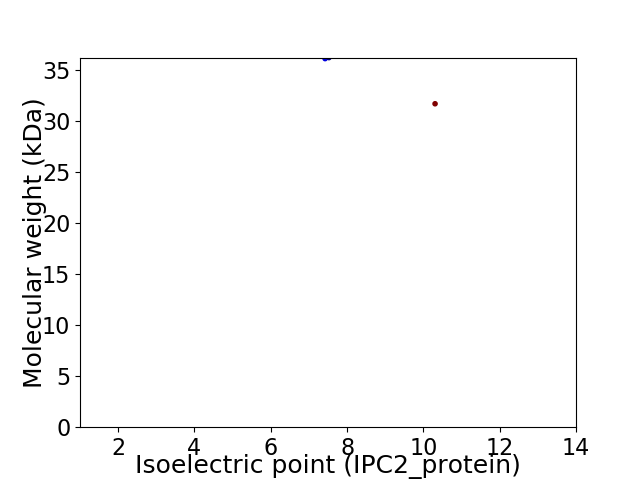
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W5U284|W5U284_9VIRU Putative capsid protein OS=Dragonfly larvae associated circular virus-9 OX=1454030 PE=4 SV=1
MM1 pKa = 7.24SARR4 pKa = 11.84VSFKK8 pKa = 10.37FSLIKK13 pKa = 9.95TSATCCSSSIPKK25 pKa = 9.24MGHH28 pKa = 4.75QVKK31 pKa = 10.07RR32 pKa = 11.84WCFTLNNYY40 pKa = 7.52TEE42 pKa = 4.29EE43 pKa = 4.34EE44 pKa = 4.11YY45 pKa = 11.19KK46 pKa = 10.92EE47 pKa = 3.88IVEE50 pKa = 4.45KK51 pKa = 10.94IEE53 pKa = 3.41WTYY56 pKa = 10.91IIIGRR61 pKa = 11.84EE62 pKa = 3.91VGEE65 pKa = 4.49EE66 pKa = 4.23GTPHH70 pKa = 5.48LQGYY74 pKa = 9.57IEE76 pKa = 4.51LKK78 pKa = 10.49KK79 pKa = 10.67KK80 pKa = 10.85SSLNQIKK87 pKa = 10.18KK88 pKa = 9.85VNQKK92 pKa = 9.13IHH94 pKa = 6.08WEE96 pKa = 3.91QLRR99 pKa = 11.84GKK101 pKa = 8.62PFQAADD107 pKa = 3.52YY108 pKa = 9.96CKK110 pKa = 10.57KK111 pKa = 10.57EE112 pKa = 4.1NNFQEE117 pKa = 4.15WGHH120 pKa = 7.76ISMSGGGTTEE130 pKa = 4.14KK131 pKa = 10.17IGAIIEE137 pKa = 4.28KK138 pKa = 9.87AKK140 pKa = 10.45SGGMRR145 pKa = 11.84EE146 pKa = 4.17VLKK149 pKa = 10.72DD150 pKa = 3.65PPKK153 pKa = 10.37LATVRR158 pKa = 11.84LIEE161 pKa = 4.98KK162 pKa = 9.39YY163 pKa = 10.42LSYY166 pKa = 11.04CEE168 pKa = 4.38EE169 pKa = 3.83EE170 pKa = 4.67RR171 pKa = 11.84KK172 pKa = 9.86EE173 pKa = 3.93KK174 pKa = 10.52PYY176 pKa = 10.77VHH178 pKa = 7.1WNLGNITTYY187 pKa = 9.07TGRR190 pKa = 11.84MTPNGGDD197 pKa = 3.46GYY199 pKa = 11.02DD200 pKa = 3.43KK201 pKa = 11.55NEE203 pKa = 4.5TIVIDD208 pKa = 4.11DD209 pKa = 4.06FRR211 pKa = 11.84ASIMRR216 pKa = 11.84FTYY219 pKa = 10.71LLRR222 pKa = 11.84ILDD225 pKa = 4.64RR226 pKa = 11.84YY227 pKa = 8.78PMRR230 pKa = 11.84VEE232 pKa = 3.94VKK234 pKa = 10.23GGYY237 pKa = 8.97RR238 pKa = 11.84QLNSKK243 pKa = 10.51NIVITSIIHH252 pKa = 6.41PEE254 pKa = 3.87SCYY257 pKa = 10.82HH258 pKa = 6.11MEE260 pKa = 4.33NEE262 pKa = 5.26PIRR265 pKa = 11.84QLMRR269 pKa = 11.84RR270 pKa = 11.84IDD272 pKa = 4.9KK273 pKa = 9.14ITEE276 pKa = 4.16TNNKK280 pKa = 9.81NILNDD285 pKa = 3.31VMLQATSEE293 pKa = 4.17EE294 pKa = 4.86SVGNTNLHH302 pKa = 6.55FLICDD307 pKa = 3.65EE308 pKa = 4.62EE309 pKa = 5.21LGEE312 pKa = 4.19
MM1 pKa = 7.24SARR4 pKa = 11.84VSFKK8 pKa = 10.37FSLIKK13 pKa = 9.95TSATCCSSSIPKK25 pKa = 9.24MGHH28 pKa = 4.75QVKK31 pKa = 10.07RR32 pKa = 11.84WCFTLNNYY40 pKa = 7.52TEE42 pKa = 4.29EE43 pKa = 4.34EE44 pKa = 4.11YY45 pKa = 11.19KK46 pKa = 10.92EE47 pKa = 3.88IVEE50 pKa = 4.45KK51 pKa = 10.94IEE53 pKa = 3.41WTYY56 pKa = 10.91IIIGRR61 pKa = 11.84EE62 pKa = 3.91VGEE65 pKa = 4.49EE66 pKa = 4.23GTPHH70 pKa = 5.48LQGYY74 pKa = 9.57IEE76 pKa = 4.51LKK78 pKa = 10.49KK79 pKa = 10.67KK80 pKa = 10.85SSLNQIKK87 pKa = 10.18KK88 pKa = 9.85VNQKK92 pKa = 9.13IHH94 pKa = 6.08WEE96 pKa = 3.91QLRR99 pKa = 11.84GKK101 pKa = 8.62PFQAADD107 pKa = 3.52YY108 pKa = 9.96CKK110 pKa = 10.57KK111 pKa = 10.57EE112 pKa = 4.1NNFQEE117 pKa = 4.15WGHH120 pKa = 7.76ISMSGGGTTEE130 pKa = 4.14KK131 pKa = 10.17IGAIIEE137 pKa = 4.28KK138 pKa = 9.87AKK140 pKa = 10.45SGGMRR145 pKa = 11.84EE146 pKa = 4.17VLKK149 pKa = 10.72DD150 pKa = 3.65PPKK153 pKa = 10.37LATVRR158 pKa = 11.84LIEE161 pKa = 4.98KK162 pKa = 9.39YY163 pKa = 10.42LSYY166 pKa = 11.04CEE168 pKa = 4.38EE169 pKa = 3.83EE170 pKa = 4.67RR171 pKa = 11.84KK172 pKa = 9.86EE173 pKa = 3.93KK174 pKa = 10.52PYY176 pKa = 10.77VHH178 pKa = 7.1WNLGNITTYY187 pKa = 9.07TGRR190 pKa = 11.84MTPNGGDD197 pKa = 3.46GYY199 pKa = 11.02DD200 pKa = 3.43KK201 pKa = 11.55NEE203 pKa = 4.5TIVIDD208 pKa = 4.11DD209 pKa = 4.06FRR211 pKa = 11.84ASIMRR216 pKa = 11.84FTYY219 pKa = 10.71LLRR222 pKa = 11.84ILDD225 pKa = 4.64RR226 pKa = 11.84YY227 pKa = 8.78PMRR230 pKa = 11.84VEE232 pKa = 3.94VKK234 pKa = 10.23GGYY237 pKa = 8.97RR238 pKa = 11.84QLNSKK243 pKa = 10.51NIVITSIIHH252 pKa = 6.41PEE254 pKa = 3.87SCYY257 pKa = 10.82HH258 pKa = 6.11MEE260 pKa = 4.33NEE262 pKa = 5.26PIRR265 pKa = 11.84QLMRR269 pKa = 11.84RR270 pKa = 11.84IDD272 pKa = 4.9KK273 pKa = 9.14ITEE276 pKa = 4.16TNNKK280 pKa = 9.81NILNDD285 pKa = 3.31VMLQATSEE293 pKa = 4.17EE294 pKa = 4.86SVGNTNLHH302 pKa = 6.55FLICDD307 pKa = 3.65EE308 pKa = 4.62EE309 pKa = 5.21LGEE312 pKa = 4.19
Molecular weight: 36.11 kDa
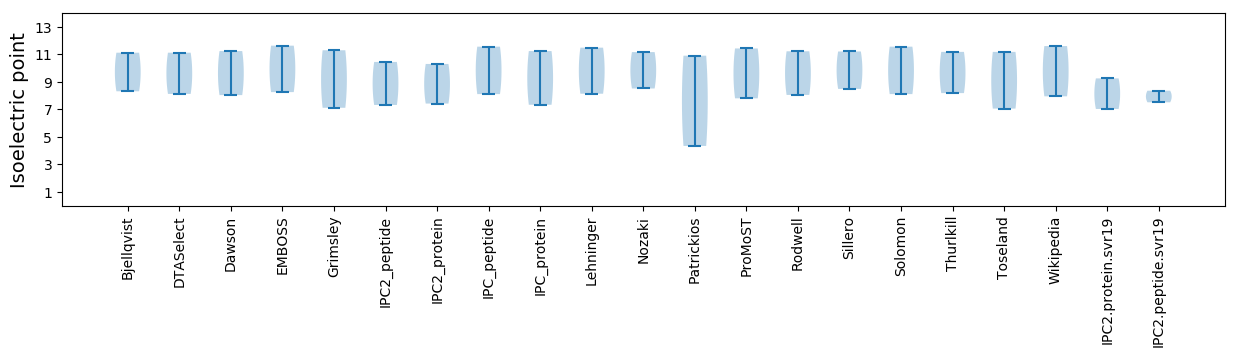
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W5U284|W5U284_9VIRU Putative capsid protein OS=Dragonfly larvae associated circular virus-9 OX=1454030 PE=4 SV=1
MM1 pKa = 6.93SHH3 pKa = 5.58FRR5 pKa = 11.84YY6 pKa = 9.54RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84ATRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84FYY17 pKa = 9.97KK18 pKa = 10.39RR19 pKa = 11.84KK20 pKa = 7.96FKK22 pKa = 10.7RR23 pKa = 11.84YY24 pKa = 5.45TRR26 pKa = 11.84RR27 pKa = 11.84HH28 pKa = 4.46ARR30 pKa = 11.84RR31 pKa = 11.84PLSRR35 pKa = 11.84PYY37 pKa = 10.0RR38 pKa = 11.84RR39 pKa = 11.84AFFKK43 pKa = 10.73CRR45 pKa = 11.84ASYY48 pKa = 9.46EE49 pKa = 4.45VKK51 pKa = 10.58GSNGPAFVAYY61 pKa = 9.16TDD63 pKa = 3.6NPVNIGDD70 pKa = 3.97FNSIQNLYY78 pKa = 8.77AQYY81 pKa = 10.39RR82 pKa = 11.84VAGMQLRR89 pKa = 11.84WIPSFNKK96 pKa = 9.8NDD98 pKa = 4.22MIPVEE103 pKa = 4.03QGEE106 pKa = 4.28QLGEE110 pKa = 3.82WLLIKK115 pKa = 10.36QDD117 pKa = 4.45DD118 pKa = 4.25ALTDD122 pKa = 3.43KK123 pKa = 11.14AFPNTVQNYY132 pKa = 8.55LNRR135 pKa = 11.84DD136 pKa = 2.98NGLRR140 pKa = 11.84RR141 pKa = 11.84RR142 pKa = 11.84PLVRR146 pKa = 11.84SWSIYY151 pKa = 9.83FKK153 pKa = 9.67MSKK156 pKa = 9.02MVPLNVASVNVVNLRR171 pKa = 11.84GGYY174 pKa = 9.62ISTVQPTATQTVRR187 pKa = 11.84AMLSSIPVNAGEE199 pKa = 4.11VLLGTFHH206 pKa = 5.43VTWYY210 pKa = 9.59VSAMQRR216 pKa = 11.84RR217 pKa = 11.84XIKK220 pKa = 10.36YY221 pKa = 9.93IIYY224 pKa = 9.66LKK226 pKa = 10.58KK227 pKa = 10.12KK228 pKa = 9.7IIFIVYY234 pKa = 8.5YY235 pKa = 8.64INFYY239 pKa = 10.48IRR241 pKa = 11.84TLIRR245 pKa = 11.84GEE247 pKa = 4.06RR248 pKa = 11.84EE249 pKa = 3.34RR250 pKa = 11.84AAVPGVSRR258 pKa = 11.84HH259 pKa = 5.07FLQILRR265 pKa = 11.84NFFF268 pKa = 3.84
MM1 pKa = 6.93SHH3 pKa = 5.58FRR5 pKa = 11.84YY6 pKa = 9.54RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84ATRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84FYY17 pKa = 9.97KK18 pKa = 10.39RR19 pKa = 11.84KK20 pKa = 7.96FKK22 pKa = 10.7RR23 pKa = 11.84YY24 pKa = 5.45TRR26 pKa = 11.84RR27 pKa = 11.84HH28 pKa = 4.46ARR30 pKa = 11.84RR31 pKa = 11.84PLSRR35 pKa = 11.84PYY37 pKa = 10.0RR38 pKa = 11.84RR39 pKa = 11.84AFFKK43 pKa = 10.73CRR45 pKa = 11.84ASYY48 pKa = 9.46EE49 pKa = 4.45VKK51 pKa = 10.58GSNGPAFVAYY61 pKa = 9.16TDD63 pKa = 3.6NPVNIGDD70 pKa = 3.97FNSIQNLYY78 pKa = 8.77AQYY81 pKa = 10.39RR82 pKa = 11.84VAGMQLRR89 pKa = 11.84WIPSFNKK96 pKa = 9.8NDD98 pKa = 4.22MIPVEE103 pKa = 4.03QGEE106 pKa = 4.28QLGEE110 pKa = 3.82WLLIKK115 pKa = 10.36QDD117 pKa = 4.45DD118 pKa = 4.25ALTDD122 pKa = 3.43KK123 pKa = 11.14AFPNTVQNYY132 pKa = 8.55LNRR135 pKa = 11.84DD136 pKa = 2.98NGLRR140 pKa = 11.84RR141 pKa = 11.84RR142 pKa = 11.84PLVRR146 pKa = 11.84SWSIYY151 pKa = 9.83FKK153 pKa = 9.67MSKK156 pKa = 9.02MVPLNVASVNVVNLRR171 pKa = 11.84GGYY174 pKa = 9.62ISTVQPTATQTVRR187 pKa = 11.84AMLSSIPVNAGEE199 pKa = 4.11VLLGTFHH206 pKa = 5.43VTWYY210 pKa = 9.59VSAMQRR216 pKa = 11.84RR217 pKa = 11.84XIKK220 pKa = 10.36YY221 pKa = 9.93IIYY224 pKa = 9.66LKK226 pKa = 10.58KK227 pKa = 10.12KK228 pKa = 9.7IIFIVYY234 pKa = 8.5YY235 pKa = 8.64INFYY239 pKa = 10.48IRR241 pKa = 11.84TLIRR245 pKa = 11.84GEE247 pKa = 4.06RR248 pKa = 11.84EE249 pKa = 3.34RR250 pKa = 11.84AAVPGVSRR258 pKa = 11.84HH259 pKa = 5.07FLQILRR265 pKa = 11.84NFFF268 pKa = 3.84
Molecular weight: 31.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
580 |
268 |
312 |
290.0 |
33.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.483 ± 1.243 | 1.379 ± 0.672 |
2.931 ± 0.213 | 6.724 ± 2.748 |
4.138 ± 1.224 | 6.034 ± 0.791 |
2.069 ± 0.385 | 8.103 ± 0.927 |
7.241 ± 1.348 | 6.897 ± 0.129 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.931 ± 0.213 | 6.207 ± 0.091 |
3.793 ± 0.457 | 3.621 ± 0.323 |
8.966 ± 2.736 | 5.862 ± 0.177 |
5.517 ± 0.695 | 6.034 ± 1.204 |
1.552 ± 0.04 | 5.345 ± 0.667 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
