
Azoarcus sp. KH32C
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Rhodocyclales; Zoogloeaceae; Azoarcus; unclassified Azoarcus
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
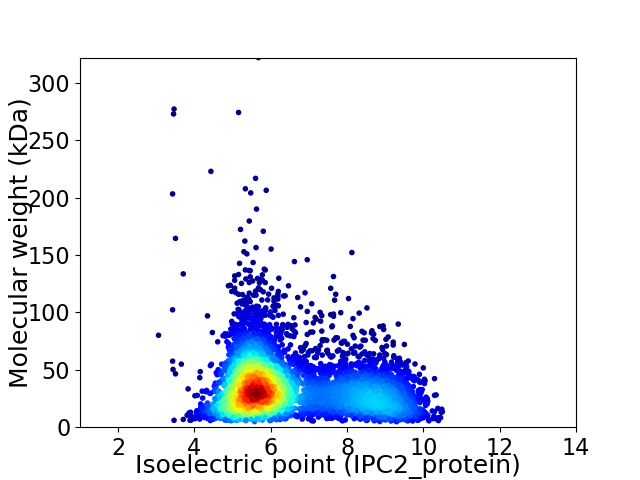
Virtual 2D-PAGE plot for 5176 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
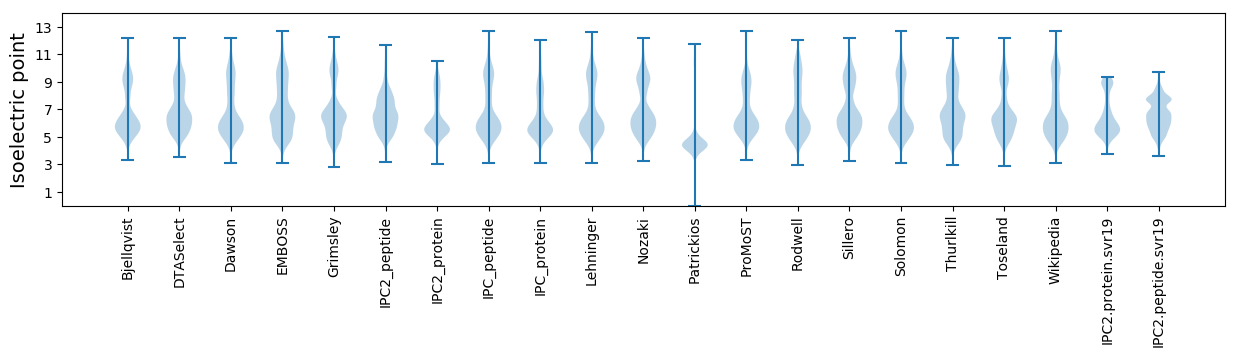
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H0Q3D0|H0Q3D0_9RHOO NAD(P)H dehydrogenase (quinone) OS=Azoarcus sp. KH32C OX=748247 GN=wrbA PE=3 SV=1
MM1 pKa = 7.0NTTAQNLAVLAAMLGLCVSSAYY23 pKa = 10.08AASAPDD29 pKa = 3.18QGGSARR35 pKa = 11.84ISGVTVEE42 pKa = 4.99SPLARR47 pKa = 11.84KK48 pKa = 9.05GADD51 pKa = 4.08DD52 pKa = 4.66PPPAPGCDD60 pKa = 3.24DD61 pKa = 4.67HH62 pKa = 7.07GTDD65 pKa = 4.07LCQSEE70 pKa = 4.37LAKK73 pKa = 11.03NGADD77 pKa = 4.46DD78 pKa = 4.98PMPQPGCDD86 pKa = 3.22DD87 pKa = 4.52HH88 pKa = 6.99GTDD91 pKa = 4.98LCLTPTAIEE100 pKa = 4.14TT101 pKa = 3.82
MM1 pKa = 7.0NTTAQNLAVLAAMLGLCVSSAYY23 pKa = 10.08AASAPDD29 pKa = 3.18QGGSARR35 pKa = 11.84ISGVTVEE42 pKa = 4.99SPLARR47 pKa = 11.84KK48 pKa = 9.05GADD51 pKa = 4.08DD52 pKa = 4.66PPPAPGCDD60 pKa = 3.24DD61 pKa = 4.67HH62 pKa = 7.07GTDD65 pKa = 4.07LCQSEE70 pKa = 4.37LAKK73 pKa = 11.03NGADD77 pKa = 4.46DD78 pKa = 4.98PMPQPGCDD86 pKa = 3.22DD87 pKa = 4.52HH88 pKa = 6.99GTDD91 pKa = 4.98LCLTPTAIEE100 pKa = 4.14TT101 pKa = 3.82
Molecular weight: 10.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H0Q037|H0Q037_9RHOO 2-isopropylmalate synthase OS=Azoarcus sp. KH32C OX=748247 GN=leuA PE=3 SV=1
MM1 pKa = 6.96NTTTVGVDD9 pKa = 3.13LAKK12 pKa = 10.79NVFVTCVADD21 pKa = 3.62GAGRR25 pKa = 11.84VIEE28 pKa = 3.98TRR30 pKa = 11.84EE31 pKa = 3.88FNRR34 pKa = 11.84SGFLAWLSTLPRR46 pKa = 11.84GTLVGMEE53 pKa = 4.2ACGGAHH59 pKa = 5.79CWGRR63 pKa = 11.84TMQSLGLEE71 pKa = 4.28PRR73 pKa = 11.84LMAPEE78 pKa = 4.1FVRR81 pKa = 11.84PYY83 pKa = 10.51RR84 pKa = 11.84KK85 pKa = 9.6RR86 pKa = 11.84QAVKK90 pKa = 10.39NDD92 pKa = 3.28RR93 pKa = 11.84ADD95 pKa = 3.18AVAIVAALLAPGMRR109 pKa = 11.84FIPVKK114 pKa = 10.29TEE116 pKa = 3.57AQQQRR121 pKa = 11.84LAWHH125 pKa = 6.11SLRR128 pKa = 11.84LGWIEE133 pKa = 4.64EE134 pKa = 4.06RR135 pKa = 11.84TALLNRR141 pKa = 11.84IRR143 pKa = 11.84GLLAEE148 pKa = 4.99FGMVVDD154 pKa = 4.5TGAMRR159 pKa = 11.84LRR161 pKa = 11.84RR162 pKa = 11.84KK163 pKa = 9.46LAEE166 pKa = 4.31HH167 pKa = 6.67EE168 pKa = 4.27FDD170 pKa = 3.6TAQPAPIRR178 pKa = 11.84QLIQGVRR185 pKa = 11.84DD186 pKa = 3.45QLDD189 pKa = 3.44ALDD192 pKa = 4.83ARR194 pKa = 11.84IAEE197 pKa = 4.77CDD199 pKa = 3.41RR200 pKa = 11.84QIATQQADD208 pKa = 3.31DD209 pKa = 3.5AAARR213 pKa = 11.84RR214 pKa = 11.84VRR216 pKa = 11.84DD217 pKa = 3.58LPGVGLLTADD227 pKa = 3.45AVVASVGDD235 pKa = 3.23ATMFHH240 pKa = 7.03NGRR243 pKa = 11.84QFAAWLGLTPSQYY256 pKa = 11.14SSGGKK261 pKa = 8.98PKK263 pKa = 10.47LGRR266 pKa = 11.84ITRR269 pKa = 11.84RR270 pKa = 11.84GDD272 pKa = 3.07DD273 pKa = 3.56YY274 pKa = 11.76LRR276 pKa = 11.84TLLVQGARR284 pKa = 11.84SVLAAALRR292 pKa = 11.84KK293 pKa = 9.52ARR295 pKa = 11.84NTRR298 pKa = 11.84EE299 pKa = 3.7ALTRR303 pKa = 11.84LQQWIVTLHH312 pKa = 5.97ARR314 pKa = 11.84VGYY317 pKa = 10.27HH318 pKa = 5.32KK319 pKa = 9.54TLVAIANKK327 pKa = 9.69HH328 pKa = 5.55ARR330 pKa = 11.84QLWAVLAKK338 pKa = 10.81GEE340 pKa = 4.48TYY342 pKa = 10.77NPEE345 pKa = 3.75AWRR348 pKa = 11.84QRR350 pKa = 11.84PDD352 pKa = 3.08SATEE356 pKa = 3.66PARR359 pKa = 3.8
MM1 pKa = 6.96NTTTVGVDD9 pKa = 3.13LAKK12 pKa = 10.79NVFVTCVADD21 pKa = 3.62GAGRR25 pKa = 11.84VIEE28 pKa = 3.98TRR30 pKa = 11.84EE31 pKa = 3.88FNRR34 pKa = 11.84SGFLAWLSTLPRR46 pKa = 11.84GTLVGMEE53 pKa = 4.2ACGGAHH59 pKa = 5.79CWGRR63 pKa = 11.84TMQSLGLEE71 pKa = 4.28PRR73 pKa = 11.84LMAPEE78 pKa = 4.1FVRR81 pKa = 11.84PYY83 pKa = 10.51RR84 pKa = 11.84KK85 pKa = 9.6RR86 pKa = 11.84QAVKK90 pKa = 10.39NDD92 pKa = 3.28RR93 pKa = 11.84ADD95 pKa = 3.18AVAIVAALLAPGMRR109 pKa = 11.84FIPVKK114 pKa = 10.29TEE116 pKa = 3.57AQQQRR121 pKa = 11.84LAWHH125 pKa = 6.11SLRR128 pKa = 11.84LGWIEE133 pKa = 4.64EE134 pKa = 4.06RR135 pKa = 11.84TALLNRR141 pKa = 11.84IRR143 pKa = 11.84GLLAEE148 pKa = 4.99FGMVVDD154 pKa = 4.5TGAMRR159 pKa = 11.84LRR161 pKa = 11.84RR162 pKa = 11.84KK163 pKa = 9.46LAEE166 pKa = 4.31HH167 pKa = 6.67EE168 pKa = 4.27FDD170 pKa = 3.6TAQPAPIRR178 pKa = 11.84QLIQGVRR185 pKa = 11.84DD186 pKa = 3.45QLDD189 pKa = 3.44ALDD192 pKa = 4.83ARR194 pKa = 11.84IAEE197 pKa = 4.77CDD199 pKa = 3.41RR200 pKa = 11.84QIATQQADD208 pKa = 3.31DD209 pKa = 3.5AAARR213 pKa = 11.84RR214 pKa = 11.84VRR216 pKa = 11.84DD217 pKa = 3.58LPGVGLLTADD227 pKa = 3.45AVVASVGDD235 pKa = 3.23ATMFHH240 pKa = 7.03NGRR243 pKa = 11.84QFAAWLGLTPSQYY256 pKa = 11.14SSGGKK261 pKa = 8.98PKK263 pKa = 10.47LGRR266 pKa = 11.84ITRR269 pKa = 11.84RR270 pKa = 11.84GDD272 pKa = 3.07DD273 pKa = 3.56YY274 pKa = 11.76LRR276 pKa = 11.84TLLVQGARR284 pKa = 11.84SVLAAALRR292 pKa = 11.84KK293 pKa = 9.52ARR295 pKa = 11.84NTRR298 pKa = 11.84EE299 pKa = 3.7ALTRR303 pKa = 11.84LQQWIVTLHH312 pKa = 5.97ARR314 pKa = 11.84VGYY317 pKa = 10.27HH318 pKa = 5.32KK319 pKa = 9.54TLVAIANKK327 pKa = 9.69HH328 pKa = 5.55ARR330 pKa = 11.84QLWAVLAKK338 pKa = 10.81GEE340 pKa = 4.48TYY342 pKa = 10.77NPEE345 pKa = 3.75AWRR348 pKa = 11.84QRR350 pKa = 11.84PDD352 pKa = 3.08SATEE356 pKa = 3.66PARR359 pKa = 3.8
Molecular weight: 39.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1764397 |
41 |
2915 |
340.9 |
37.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.335 ± 0.045 | 1.027 ± 0.012 |
5.57 ± 0.027 | 5.979 ± 0.03 |
3.644 ± 0.022 | 8.367 ± 0.036 |
2.255 ± 0.016 | 4.757 ± 0.024 |
3.124 ± 0.027 | 10.537 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.387 ± 0.015 | 2.638 ± 0.018 |
5.059 ± 0.024 | 3.325 ± 0.021 |
7.415 ± 0.038 | 5.252 ± 0.023 |
5.025 ± 0.033 | 7.675 ± 0.03 |
1.351 ± 0.013 | 2.279 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
