
Beihai weivirus-like virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.55
Get precalculated fractions of proteins
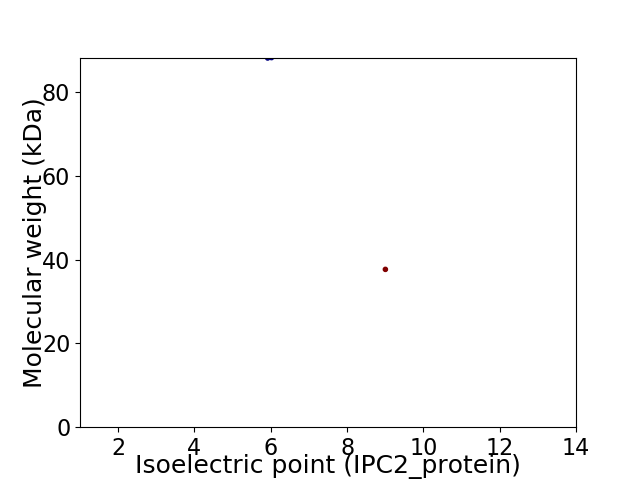
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
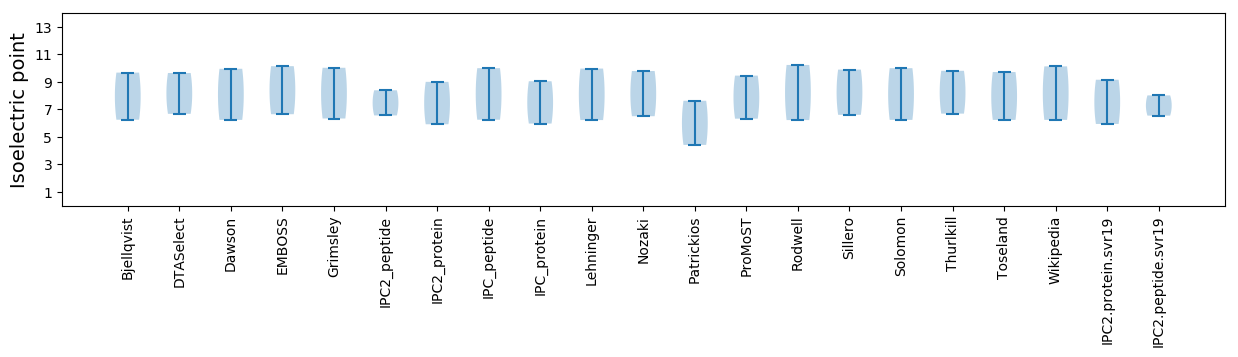
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KLC2|A0A1L3KLC2_9VIRU Uncharacterized protein OS=Beihai weivirus-like virus 4 OX=1922752 PE=4 SV=1
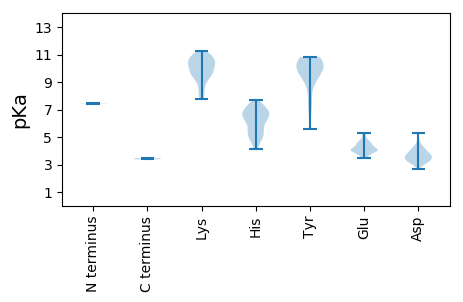
MM1 pKa = 7.44LALRR5 pKa = 11.84LGQTDD10 pKa = 3.69RR11 pKa = 11.84QIGEE15 pKa = 4.02VAPFLLRR22 pKa = 11.84HH23 pKa = 4.98AAGRR27 pKa = 11.84QGHH30 pKa = 5.21WRR32 pKa = 11.84ITAQGYY38 pKa = 7.54DD39 pKa = 3.37HH40 pKa = 7.68PAAQSTKK47 pKa = 9.57AYY49 pKa = 9.27SVVYY53 pKa = 9.96TEE55 pKa = 5.04EE56 pKa = 4.21EE57 pKa = 3.95FSKK60 pKa = 10.69LSRR63 pKa = 11.84VVYY66 pKa = 10.29HH67 pKa = 6.5GLRR70 pKa = 11.84NSTKK74 pKa = 8.63PTSLHH79 pKa = 6.52ASLEE83 pKa = 4.22NVAQVLWPTGSPDD96 pKa = 3.32HH97 pKa = 6.3VAMRR101 pKa = 11.84HH102 pKa = 4.55VGVFVMMQYY111 pKa = 7.27MTSDD115 pKa = 3.33EE116 pKa = 4.38TAVRR120 pKa = 11.84PLARR124 pKa = 11.84WAMVAGVGIGLMVRR138 pKa = 11.84SVADD142 pKa = 3.52GWKK145 pKa = 9.77LATIAGLSPVLLARR159 pKa = 11.84LVSRR163 pKa = 11.84GRR165 pKa = 11.84AIPILEE171 pKa = 3.98RR172 pKa = 11.84VGASQAAVAPNPGGSGPPPQAAPAPPPPPQGEE204 pKa = 4.18PPPPGDD210 pKa = 3.62AEE212 pKa = 4.05TDD214 pKa = 3.02EE215 pKa = 4.46GVRR218 pKa = 11.84VEE220 pKa = 4.66EE221 pKa = 4.54LNEE224 pKa = 4.03EE225 pKa = 4.31PPVIMDD231 pKa = 4.25TPALPDD237 pKa = 3.93DD238 pKa = 4.54AATAAAASNDD248 pKa = 3.27RR249 pKa = 11.84RR250 pKa = 11.84TMQVGDD256 pKa = 3.72VVAVIGQAFNKK267 pKa = 10.1DD268 pKa = 3.24EE269 pKa = 4.81PVNHH273 pKa = 6.04MPVVGCLVGPCQVKK287 pKa = 9.9PNVYY291 pKa = 10.71AKK293 pKa = 9.47TASNLKK299 pKa = 10.03AAIEE303 pKa = 3.82EE304 pKa = 4.64RR305 pKa = 11.84ITKK308 pKa = 10.09KK309 pKa = 10.42ARR311 pKa = 11.84KK312 pKa = 8.42CQLSKK317 pKa = 10.63KK318 pKa = 9.74DD319 pKa = 3.26KK320 pKa = 10.75ARR322 pKa = 11.84IGGLVRR328 pKa = 11.84QSMSCHH334 pKa = 5.6RR335 pKa = 11.84VRR337 pKa = 11.84GVFSKK342 pKa = 10.87RR343 pKa = 11.84KK344 pKa = 7.73IEE346 pKa = 3.6AWAIEE351 pKa = 4.03NLDD354 pKa = 3.86LEE356 pKa = 5.28LIRR359 pKa = 11.84SGKK362 pKa = 8.94WSVEE366 pKa = 3.67RR367 pKa = 11.84FRR369 pKa = 11.84ASLEE373 pKa = 3.58ALYY376 pKa = 10.81ARR378 pKa = 11.84EE379 pKa = 3.81YY380 pKa = 9.16PTYY383 pKa = 10.29QFKK386 pKa = 11.08AGIKK390 pKa = 7.88PEE392 pKa = 4.04CMAEE396 pKa = 4.19GKK398 pKa = 10.24APRR401 pKa = 11.84MLIADD406 pKa = 4.1GDD408 pKa = 4.23DD409 pKa = 3.83GQLMALAVVRR419 pKa = 11.84CFEE422 pKa = 4.66DD423 pKa = 3.6LLFEE427 pKa = 4.22HH428 pKa = 7.01FEE430 pKa = 4.23RR431 pKa = 11.84KK432 pKa = 9.43SIKK435 pKa = 10.43HH436 pKa = 4.13MAKK439 pKa = 9.73RR440 pKa = 11.84EE441 pKa = 3.97AMDD444 pKa = 3.49RR445 pKa = 11.84VVKK448 pKa = 10.26EE449 pKa = 4.07LSKK452 pKa = 10.95KK453 pKa = 8.46GAKK456 pKa = 9.6AVEE459 pKa = 4.46GDD461 pKa = 3.29GSAWDD466 pKa = 3.82TTCNVEE472 pKa = 4.35IRR474 pKa = 11.84ALVEE478 pKa = 4.03NPVLRR483 pKa = 11.84HH484 pKa = 5.07ICEE487 pKa = 4.22VLCGLGVVPEE497 pKa = 4.05SWLRR501 pKa = 11.84EE502 pKa = 3.66HH503 pKa = 7.25SAACEE508 pKa = 3.96QKK510 pKa = 9.31TLRR513 pKa = 11.84LFFKK517 pKa = 11.03NKK519 pKa = 8.0MEE521 pKa = 4.37SMSVTIDD528 pKa = 3.9AIRR531 pKa = 11.84RR532 pKa = 11.84SGHH535 pKa = 6.55RR536 pKa = 11.84GTSCLNWWVNFTLWVSSIFAEE557 pKa = 4.39PEE559 pKa = 3.74RR560 pKa = 11.84FLDD563 pKa = 3.56PDD565 pKa = 3.28VRR567 pKa = 11.84TGEE570 pKa = 4.5DD571 pKa = 3.0LTGQQRR577 pKa = 11.84WWNGCFEE584 pKa = 5.17GDD586 pKa = 5.31DD587 pKa = 4.28SLCTMKK593 pKa = 10.66PPMEE597 pKa = 4.64EE598 pKa = 3.82GDD600 pKa = 3.52RR601 pKa = 11.84MSEE604 pKa = 3.59IFLEE608 pKa = 4.92FWRR611 pKa = 11.84DD612 pKa = 2.68AGFNMKK618 pKa = 9.64IVFCTTRR625 pKa = 11.84ATFVGWHH632 pKa = 6.47IGCTDD637 pKa = 3.42GEE639 pKa = 4.63LNEE642 pKa = 4.39HH643 pKa = 7.02RR644 pKa = 11.84CPEE647 pKa = 4.16LPRR650 pKa = 11.84ALANSGVSVSPQGVEE665 pKa = 3.5AGRR668 pKa = 11.84DD669 pKa = 3.47GKK671 pKa = 11.25LSVVKK676 pKa = 10.36VLAAASALARR686 pKa = 11.84ASDD689 pKa = 3.83FSGILPSVSQKK700 pKa = 10.81YY701 pKa = 10.55LDD703 pKa = 3.81FAEE706 pKa = 4.75EE707 pKa = 4.46CTTSDD712 pKa = 3.72FEE714 pKa = 4.59DD715 pKa = 4.51RR716 pKa = 11.84EE717 pKa = 3.84MSYY720 pKa = 10.76RR721 pKa = 11.84AFGEE725 pKa = 4.07AGYY728 pKa = 9.93KK729 pKa = 10.6ANDD732 pKa = 3.15VRR734 pKa = 11.84EE735 pKa = 4.06QVQARR740 pKa = 11.84NLEE743 pKa = 4.07VTPEE747 pKa = 4.02KK748 pKa = 10.57EE749 pKa = 4.18QEE751 pKa = 3.95TLRR754 pKa = 11.84ALGYY758 pKa = 10.07QATHH762 pKa = 6.79DD763 pKa = 4.08EE764 pKa = 3.97IMTFRR769 pKa = 11.84EE770 pKa = 4.66YY771 pKa = 10.1IWSLEE776 pKa = 3.87PAALVAYY783 pKa = 10.22DD784 pKa = 3.97SFRR787 pKa = 11.84EE788 pKa = 4.01SLPPSWRR795 pKa = 11.84AAA797 pKa = 3.38
MM1 pKa = 7.44LALRR5 pKa = 11.84LGQTDD10 pKa = 3.69RR11 pKa = 11.84QIGEE15 pKa = 4.02VAPFLLRR22 pKa = 11.84HH23 pKa = 4.98AAGRR27 pKa = 11.84QGHH30 pKa = 5.21WRR32 pKa = 11.84ITAQGYY38 pKa = 7.54DD39 pKa = 3.37HH40 pKa = 7.68PAAQSTKK47 pKa = 9.57AYY49 pKa = 9.27SVVYY53 pKa = 9.96TEE55 pKa = 5.04EE56 pKa = 4.21EE57 pKa = 3.95FSKK60 pKa = 10.69LSRR63 pKa = 11.84VVYY66 pKa = 10.29HH67 pKa = 6.5GLRR70 pKa = 11.84NSTKK74 pKa = 8.63PTSLHH79 pKa = 6.52ASLEE83 pKa = 4.22NVAQVLWPTGSPDD96 pKa = 3.32HH97 pKa = 6.3VAMRR101 pKa = 11.84HH102 pKa = 4.55VGVFVMMQYY111 pKa = 7.27MTSDD115 pKa = 3.33EE116 pKa = 4.38TAVRR120 pKa = 11.84PLARR124 pKa = 11.84WAMVAGVGIGLMVRR138 pKa = 11.84SVADD142 pKa = 3.52GWKK145 pKa = 9.77LATIAGLSPVLLARR159 pKa = 11.84LVSRR163 pKa = 11.84GRR165 pKa = 11.84AIPILEE171 pKa = 3.98RR172 pKa = 11.84VGASQAAVAPNPGGSGPPPQAAPAPPPPPQGEE204 pKa = 4.18PPPPGDD210 pKa = 3.62AEE212 pKa = 4.05TDD214 pKa = 3.02EE215 pKa = 4.46GVRR218 pKa = 11.84VEE220 pKa = 4.66EE221 pKa = 4.54LNEE224 pKa = 4.03EE225 pKa = 4.31PPVIMDD231 pKa = 4.25TPALPDD237 pKa = 3.93DD238 pKa = 4.54AATAAAASNDD248 pKa = 3.27RR249 pKa = 11.84RR250 pKa = 11.84TMQVGDD256 pKa = 3.72VVAVIGQAFNKK267 pKa = 10.1DD268 pKa = 3.24EE269 pKa = 4.81PVNHH273 pKa = 6.04MPVVGCLVGPCQVKK287 pKa = 9.9PNVYY291 pKa = 10.71AKK293 pKa = 9.47TASNLKK299 pKa = 10.03AAIEE303 pKa = 3.82EE304 pKa = 4.64RR305 pKa = 11.84ITKK308 pKa = 10.09KK309 pKa = 10.42ARR311 pKa = 11.84KK312 pKa = 8.42CQLSKK317 pKa = 10.63KK318 pKa = 9.74DD319 pKa = 3.26KK320 pKa = 10.75ARR322 pKa = 11.84IGGLVRR328 pKa = 11.84QSMSCHH334 pKa = 5.6RR335 pKa = 11.84VRR337 pKa = 11.84GVFSKK342 pKa = 10.87RR343 pKa = 11.84KK344 pKa = 7.73IEE346 pKa = 3.6AWAIEE351 pKa = 4.03NLDD354 pKa = 3.86LEE356 pKa = 5.28LIRR359 pKa = 11.84SGKK362 pKa = 8.94WSVEE366 pKa = 3.67RR367 pKa = 11.84FRR369 pKa = 11.84ASLEE373 pKa = 3.58ALYY376 pKa = 10.81ARR378 pKa = 11.84EE379 pKa = 3.81YY380 pKa = 9.16PTYY383 pKa = 10.29QFKK386 pKa = 11.08AGIKK390 pKa = 7.88PEE392 pKa = 4.04CMAEE396 pKa = 4.19GKK398 pKa = 10.24APRR401 pKa = 11.84MLIADD406 pKa = 4.1GDD408 pKa = 4.23DD409 pKa = 3.83GQLMALAVVRR419 pKa = 11.84CFEE422 pKa = 4.66DD423 pKa = 3.6LLFEE427 pKa = 4.22HH428 pKa = 7.01FEE430 pKa = 4.23RR431 pKa = 11.84KK432 pKa = 9.43SIKK435 pKa = 10.43HH436 pKa = 4.13MAKK439 pKa = 9.73RR440 pKa = 11.84EE441 pKa = 3.97AMDD444 pKa = 3.49RR445 pKa = 11.84VVKK448 pKa = 10.26EE449 pKa = 4.07LSKK452 pKa = 10.95KK453 pKa = 8.46GAKK456 pKa = 9.6AVEE459 pKa = 4.46GDD461 pKa = 3.29GSAWDD466 pKa = 3.82TTCNVEE472 pKa = 4.35IRR474 pKa = 11.84ALVEE478 pKa = 4.03NPVLRR483 pKa = 11.84HH484 pKa = 5.07ICEE487 pKa = 4.22VLCGLGVVPEE497 pKa = 4.05SWLRR501 pKa = 11.84EE502 pKa = 3.66HH503 pKa = 7.25SAACEE508 pKa = 3.96QKK510 pKa = 9.31TLRR513 pKa = 11.84LFFKK517 pKa = 11.03NKK519 pKa = 8.0MEE521 pKa = 4.37SMSVTIDD528 pKa = 3.9AIRR531 pKa = 11.84RR532 pKa = 11.84SGHH535 pKa = 6.55RR536 pKa = 11.84GTSCLNWWVNFTLWVSSIFAEE557 pKa = 4.39PEE559 pKa = 3.74RR560 pKa = 11.84FLDD563 pKa = 3.56PDD565 pKa = 3.28VRR567 pKa = 11.84TGEE570 pKa = 4.5DD571 pKa = 3.0LTGQQRR577 pKa = 11.84WWNGCFEE584 pKa = 5.17GDD586 pKa = 5.31DD587 pKa = 4.28SLCTMKK593 pKa = 10.66PPMEE597 pKa = 4.64EE598 pKa = 3.82GDD600 pKa = 3.52RR601 pKa = 11.84MSEE604 pKa = 3.59IFLEE608 pKa = 4.92FWRR611 pKa = 11.84DD612 pKa = 2.68AGFNMKK618 pKa = 9.64IVFCTTRR625 pKa = 11.84ATFVGWHH632 pKa = 6.47IGCTDD637 pKa = 3.42GEE639 pKa = 4.63LNEE642 pKa = 4.39HH643 pKa = 7.02RR644 pKa = 11.84CPEE647 pKa = 4.16LPRR650 pKa = 11.84ALANSGVSVSPQGVEE665 pKa = 3.5AGRR668 pKa = 11.84DD669 pKa = 3.47GKK671 pKa = 11.25LSVVKK676 pKa = 10.36VLAAASALARR686 pKa = 11.84ASDD689 pKa = 3.83FSGILPSVSQKK700 pKa = 10.81YY701 pKa = 10.55LDD703 pKa = 3.81FAEE706 pKa = 4.75EE707 pKa = 4.46CTTSDD712 pKa = 3.72FEE714 pKa = 4.59DD715 pKa = 4.51RR716 pKa = 11.84EE717 pKa = 3.84MSYY720 pKa = 10.76RR721 pKa = 11.84AFGEE725 pKa = 4.07AGYY728 pKa = 9.93KK729 pKa = 10.6ANDD732 pKa = 3.15VRR734 pKa = 11.84EE735 pKa = 4.06QVQARR740 pKa = 11.84NLEE743 pKa = 4.07VTPEE747 pKa = 4.02KK748 pKa = 10.57EE749 pKa = 4.18QEE751 pKa = 3.95TLRR754 pKa = 11.84ALGYY758 pKa = 10.07QATHH762 pKa = 6.79DD763 pKa = 4.08EE764 pKa = 3.97IMTFRR769 pKa = 11.84EE770 pKa = 4.66YY771 pKa = 10.1IWSLEE776 pKa = 3.87PAALVAYY783 pKa = 10.22DD784 pKa = 3.97SFRR787 pKa = 11.84EE788 pKa = 4.01SLPPSWRR795 pKa = 11.84AAA797 pKa = 3.38
Molecular weight: 88.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KLC2|A0A1L3KLC2_9VIRU Uncharacterized protein OS=Beihai weivirus-like virus 4 OX=1922752 PE=4 SV=1
MM1 pKa = 7.46TPNKK5 pKa = 9.1PAQRR9 pKa = 11.84NGRR12 pKa = 11.84KK13 pKa = 9.4LRR15 pKa = 11.84DD16 pKa = 3.47SGDD19 pKa = 3.16KK20 pKa = 10.46SKK22 pKa = 10.93RR23 pKa = 11.84PQRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84GGAASRR34 pKa = 11.84THH36 pKa = 6.65ANRR39 pKa = 11.84VLATGVGNVPTKK51 pKa = 10.57AWGSSRR57 pKa = 11.84GGSLLCWDD65 pKa = 4.81AKK67 pKa = 10.64HH68 pKa = 6.04PHH70 pKa = 6.55HH71 pKa = 7.04LSLPRR76 pKa = 11.84AVGPYY81 pKa = 5.61TTIRR85 pKa = 11.84ATRR88 pKa = 11.84RR89 pKa = 11.84VATGRR94 pKa = 11.84IANIIGTFKK103 pKa = 10.72SRR105 pKa = 11.84HH106 pKa = 5.19NSSSLGEE113 pKa = 4.06ASADD117 pKa = 3.98TKK119 pKa = 10.01WSEE122 pKa = 4.01IIMISDD128 pKa = 3.52HH129 pKa = 7.1VSANPINGTDD139 pKa = 3.02NTFFNNIDD147 pKa = 3.62LGEE150 pKa = 4.52LGSAATLVPSALTVQIVCPDD170 pKa = 3.2NLAADD175 pKa = 4.1GAKK178 pKa = 9.45GVVYY182 pKa = 10.35AGVMNTQAMVGGKK195 pKa = 9.42SDD197 pKa = 3.24SWDD200 pKa = 2.98AYY202 pKa = 9.0MNRR205 pKa = 11.84FVQFQSPRR213 pKa = 11.84MLAATKK219 pKa = 10.12LALRR223 pKa = 11.84GVTINSYY230 pKa = 9.59PLDD233 pKa = 3.65MSEE236 pKa = 5.15VSDD239 pKa = 3.56FTQLHH244 pKa = 5.22EE245 pKa = 4.62RR246 pKa = 11.84EE247 pKa = 4.17GLEE250 pKa = 3.96YY251 pKa = 9.92TGTMSSLSPQPAGWAPICIYY271 pKa = 10.56NPQGALLEE279 pKa = 4.02LLITVEE285 pKa = 3.69YY286 pKa = 10.09RR287 pKa = 11.84VRR289 pKa = 11.84FDD291 pKa = 4.25LEE293 pKa = 4.24HH294 pKa = 6.86PASASHH300 pKa = 5.16VHH302 pKa = 6.27HH303 pKa = 6.96PVASDD308 pKa = 3.62STWDD312 pKa = 3.27KK313 pKa = 10.14LTRR316 pKa = 11.84KK317 pKa = 9.76ASSLGHH323 pKa = 5.59GVMDD327 pKa = 4.11IADD330 pKa = 3.62VVANIGSAVRR340 pKa = 11.84TGRR343 pKa = 11.84QMLSAFGG350 pKa = 3.46
MM1 pKa = 7.46TPNKK5 pKa = 9.1PAQRR9 pKa = 11.84NGRR12 pKa = 11.84KK13 pKa = 9.4LRR15 pKa = 11.84DD16 pKa = 3.47SGDD19 pKa = 3.16KK20 pKa = 10.46SKK22 pKa = 10.93RR23 pKa = 11.84PQRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84GGAASRR34 pKa = 11.84THH36 pKa = 6.65ANRR39 pKa = 11.84VLATGVGNVPTKK51 pKa = 10.57AWGSSRR57 pKa = 11.84GGSLLCWDD65 pKa = 4.81AKK67 pKa = 10.64HH68 pKa = 6.04PHH70 pKa = 6.55HH71 pKa = 7.04LSLPRR76 pKa = 11.84AVGPYY81 pKa = 5.61TTIRR85 pKa = 11.84ATRR88 pKa = 11.84RR89 pKa = 11.84VATGRR94 pKa = 11.84IANIIGTFKK103 pKa = 10.72SRR105 pKa = 11.84HH106 pKa = 5.19NSSSLGEE113 pKa = 4.06ASADD117 pKa = 3.98TKK119 pKa = 10.01WSEE122 pKa = 4.01IIMISDD128 pKa = 3.52HH129 pKa = 7.1VSANPINGTDD139 pKa = 3.02NTFFNNIDD147 pKa = 3.62LGEE150 pKa = 4.52LGSAATLVPSALTVQIVCPDD170 pKa = 3.2NLAADD175 pKa = 4.1GAKK178 pKa = 9.45GVVYY182 pKa = 10.35AGVMNTQAMVGGKK195 pKa = 9.42SDD197 pKa = 3.24SWDD200 pKa = 2.98AYY202 pKa = 9.0MNRR205 pKa = 11.84FVQFQSPRR213 pKa = 11.84MLAATKK219 pKa = 10.12LALRR223 pKa = 11.84GVTINSYY230 pKa = 9.59PLDD233 pKa = 3.65MSEE236 pKa = 5.15VSDD239 pKa = 3.56FTQLHH244 pKa = 5.22EE245 pKa = 4.62RR246 pKa = 11.84EE247 pKa = 4.17GLEE250 pKa = 3.96YY251 pKa = 9.92TGTMSSLSPQPAGWAPICIYY271 pKa = 10.56NPQGALLEE279 pKa = 4.02LLITVEE285 pKa = 3.69YY286 pKa = 10.09RR287 pKa = 11.84VRR289 pKa = 11.84FDD291 pKa = 4.25LEE293 pKa = 4.24HH294 pKa = 6.86PASASHH300 pKa = 5.16VHH302 pKa = 6.27HH303 pKa = 6.96PVASDD308 pKa = 3.62STWDD312 pKa = 3.27KK313 pKa = 10.14LTRR316 pKa = 11.84KK317 pKa = 9.76ASSLGHH323 pKa = 5.59GVMDD327 pKa = 4.11IADD330 pKa = 3.62VVANIGSAVRR340 pKa = 11.84TGRR343 pKa = 11.84QMLSAFGG350 pKa = 3.46
Molecular weight: 37.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1147 |
350 |
797 |
573.5 |
62.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.811 ± 0.133 | 1.744 ± 0.492 |
4.969 ± 0.096 | 6.452 ± 1.993 |
3.051 ± 0.425 | 7.672 ± 0.657 |
2.528 ± 0.499 | 3.836 ± 0.408 |
4.359 ± 0.358 | 7.759 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.964 ± 0.059 | 3.313 ± 0.856 |
5.754 ± 0.339 | 3.139 ± 0.156 |
7.323 ± 0.1 | 7.411 ± 1.277 |
5.231 ± 0.902 | 7.759 ± 0.5 |
2.005 ± 0.161 | 1.918 ± 0.045 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
