
Sierra dome spider associated circular virus 2
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.98
Get precalculated fractions of proteins
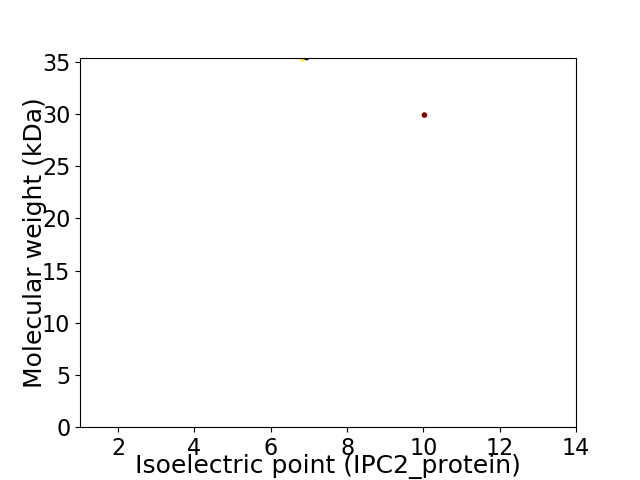
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BPD5|A0A346BPD5_9VIRU Putative capsid protein OS=Sierra dome spider associated circular virus 2 OX=2293300 PE=4 SV=1
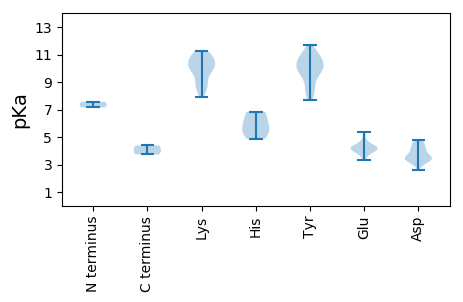
MM1 pKa = 7.56CLDD4 pKa = 4.71LLTVSNMSRR13 pKa = 11.84CRR15 pKa = 11.84NYY17 pKa = 10.3VFTLNNPQGTPMFVPDD33 pKa = 3.14KK34 pKa = 8.99MKK36 pKa = 10.95YY37 pKa = 8.59MVYY40 pKa = 10.1QKK42 pKa = 10.93EE43 pKa = 4.25SGEE46 pKa = 4.18NGTNHH51 pKa = 4.87YY52 pKa = 9.79QGYY55 pKa = 9.53VEE57 pKa = 5.06LLSPTGMPSVKK68 pKa = 10.19RR69 pKa = 11.84MIGQNPHH76 pKa = 6.43LEE78 pKa = 4.22PRR80 pKa = 11.84RR81 pKa = 11.84GTQAEE86 pKa = 3.78ARR88 pKa = 11.84AYY90 pKa = 10.29CMKK93 pKa = 10.5PEE95 pKa = 3.97TRR97 pKa = 11.84VDD99 pKa = 4.19GPWEE103 pKa = 3.9MGTFTEE109 pKa = 4.17NNQGKK114 pKa = 7.88RR115 pKa = 11.84TDD117 pKa = 4.03LEE119 pKa = 4.26LAIQDD124 pKa = 3.41IDD126 pKa = 4.17GGSSMARR133 pKa = 11.84VALDD137 pKa = 3.36HH138 pKa = 6.82PVTFVKK144 pKa = 10.58YY145 pKa = 9.76PRR147 pKa = 11.84GLSAYY152 pKa = 10.18ANMRR156 pKa = 11.84NFRR159 pKa = 11.84GTTYY163 pKa = 9.87TKK165 pKa = 10.28KK166 pKa = 10.57QSIVLWGSPGCGKK179 pKa = 9.18TSTLVNYY186 pKa = 9.62FPDD189 pKa = 4.32MYY191 pKa = 11.72VKK193 pKa = 10.1MCDD196 pKa = 3.36NKK198 pKa = 9.88WWDD201 pKa = 3.84GYY203 pKa = 11.31SDD205 pKa = 3.67QKK207 pKa = 11.23QVLLDD212 pKa = 4.47DD213 pKa = 4.8FPNTYY218 pKa = 7.96TAKK221 pKa = 8.63QTKK224 pKa = 9.93DD225 pKa = 3.27LLGEE229 pKa = 4.21LDD231 pKa = 4.59LPRR234 pKa = 11.84EE235 pKa = 4.35SKK237 pKa = 10.92GQVIKK242 pKa = 10.7PRR244 pKa = 11.84VEE246 pKa = 4.04FFFITSNFHH255 pKa = 5.53PDD257 pKa = 2.63EE258 pKa = 4.28WFEE261 pKa = 4.29SAKK264 pKa = 10.18QQDD267 pKa = 3.51RR268 pKa = 11.84LAVRR272 pKa = 11.84RR273 pKa = 11.84RR274 pKa = 11.84ITKK277 pKa = 8.85TYY279 pKa = 9.51NFDD282 pKa = 3.4VEE284 pKa = 4.18EE285 pKa = 4.68EE286 pKa = 3.32IWEE289 pKa = 4.34FVRR292 pKa = 11.84EE293 pKa = 3.95FSPYY297 pKa = 9.78NVRR300 pKa = 11.84TQEE303 pKa = 3.88LL304 pKa = 3.73
MM1 pKa = 7.56CLDD4 pKa = 4.71LLTVSNMSRR13 pKa = 11.84CRR15 pKa = 11.84NYY17 pKa = 10.3VFTLNNPQGTPMFVPDD33 pKa = 3.14KK34 pKa = 8.99MKK36 pKa = 10.95YY37 pKa = 8.59MVYY40 pKa = 10.1QKK42 pKa = 10.93EE43 pKa = 4.25SGEE46 pKa = 4.18NGTNHH51 pKa = 4.87YY52 pKa = 9.79QGYY55 pKa = 9.53VEE57 pKa = 5.06LLSPTGMPSVKK68 pKa = 10.19RR69 pKa = 11.84MIGQNPHH76 pKa = 6.43LEE78 pKa = 4.22PRR80 pKa = 11.84RR81 pKa = 11.84GTQAEE86 pKa = 3.78ARR88 pKa = 11.84AYY90 pKa = 10.29CMKK93 pKa = 10.5PEE95 pKa = 3.97TRR97 pKa = 11.84VDD99 pKa = 4.19GPWEE103 pKa = 3.9MGTFTEE109 pKa = 4.17NNQGKK114 pKa = 7.88RR115 pKa = 11.84TDD117 pKa = 4.03LEE119 pKa = 4.26LAIQDD124 pKa = 3.41IDD126 pKa = 4.17GGSSMARR133 pKa = 11.84VALDD137 pKa = 3.36HH138 pKa = 6.82PVTFVKK144 pKa = 10.58YY145 pKa = 9.76PRR147 pKa = 11.84GLSAYY152 pKa = 10.18ANMRR156 pKa = 11.84NFRR159 pKa = 11.84GTTYY163 pKa = 9.87TKK165 pKa = 10.28KK166 pKa = 10.57QSIVLWGSPGCGKK179 pKa = 9.18TSTLVNYY186 pKa = 9.62FPDD189 pKa = 4.32MYY191 pKa = 11.72VKK193 pKa = 10.1MCDD196 pKa = 3.36NKK198 pKa = 9.88WWDD201 pKa = 3.84GYY203 pKa = 11.31SDD205 pKa = 3.67QKK207 pKa = 11.23QVLLDD212 pKa = 4.47DD213 pKa = 4.8FPNTYY218 pKa = 7.96TAKK221 pKa = 8.63QTKK224 pKa = 9.93DD225 pKa = 3.27LLGEE229 pKa = 4.21LDD231 pKa = 4.59LPRR234 pKa = 11.84EE235 pKa = 4.35SKK237 pKa = 10.92GQVIKK242 pKa = 10.7PRR244 pKa = 11.84VEE246 pKa = 4.04FFFITSNFHH255 pKa = 5.53PDD257 pKa = 2.63EE258 pKa = 4.28WFEE261 pKa = 4.29SAKK264 pKa = 10.18QQDD267 pKa = 3.51RR268 pKa = 11.84LAVRR272 pKa = 11.84RR273 pKa = 11.84RR274 pKa = 11.84ITKK277 pKa = 8.85TYY279 pKa = 9.51NFDD282 pKa = 3.4VEE284 pKa = 4.18EE285 pKa = 4.68EE286 pKa = 3.32IWEE289 pKa = 4.34FVRR292 pKa = 11.84EE293 pKa = 3.95FSPYY297 pKa = 9.78NVRR300 pKa = 11.84TQEE303 pKa = 3.88LL304 pKa = 3.73
Molecular weight: 35.29 kDa
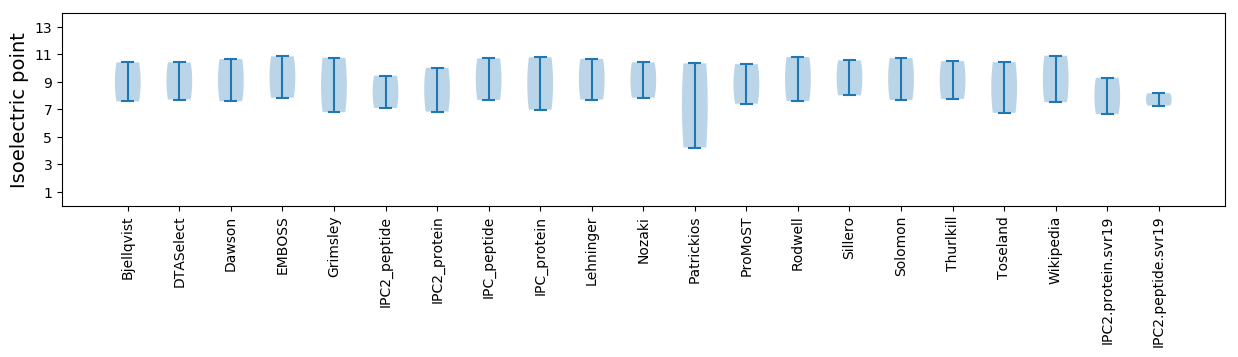
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BPD5|A0A346BPD5_9VIRU Putative capsid protein OS=Sierra dome spider associated circular virus 2 OX=2293300 PE=4 SV=1
MM1 pKa = 7.16YY2 pKa = 10.36GRR4 pKa = 11.84RR5 pKa = 11.84SYY7 pKa = 10.89RR8 pKa = 11.84RR9 pKa = 11.84YY10 pKa = 7.66RR11 pKa = 11.84TYY13 pKa = 10.96RR14 pKa = 11.84KK15 pKa = 8.91GSGRR19 pKa = 11.84ARR21 pKa = 11.84YY22 pKa = 8.39GATKK26 pKa = 9.64ARR28 pKa = 11.84RR29 pKa = 11.84VVRR32 pKa = 11.84YY33 pKa = 9.06RR34 pKa = 11.84GARR37 pKa = 11.84ARR39 pKa = 11.84GIKK42 pKa = 10.09RR43 pKa = 11.84IVNRR47 pKa = 11.84QIEE50 pKa = 4.61TKK52 pKa = 9.76EE53 pKa = 4.05YY54 pKa = 10.14TNGTQTSMLQPTQLGAFGAGYY75 pKa = 10.48NGFSDD80 pKa = 3.59TVSYY84 pKa = 8.6FKK86 pKa = 11.11PMDD89 pKa = 3.82YY90 pKa = 10.73LSQGTSRR97 pKa = 11.84NTRR100 pKa = 11.84IGDD103 pKa = 3.39AVTIMSIEE111 pKa = 3.53WRR113 pKa = 11.84YY114 pKa = 10.72CITFTDD120 pKa = 4.24YY121 pKa = 9.8EE122 pKa = 4.33QEE124 pKa = 4.58FIRR127 pKa = 11.84VMAFTAPFGFTPTYY141 pKa = 11.07GNIFEE146 pKa = 5.4APDD149 pKa = 3.51GNVDD153 pKa = 3.54VRR155 pKa = 11.84VARR158 pKa = 11.84PNTNITVLKK167 pKa = 9.91QYY169 pKa = 11.0KK170 pKa = 8.39HH171 pKa = 5.58TLSPLNSATASSSSTVNTPQVFRR194 pKa = 11.84TGCLRR199 pKa = 11.84LLFPGGRR206 pKa = 11.84KK207 pKa = 8.43FQYY210 pKa = 10.67SFNNYY215 pKa = 8.42TGSTALFNRR224 pKa = 11.84NRR226 pKa = 11.84DD227 pKa = 3.2IFIVAVPFAAKK238 pKa = 10.14RR239 pKa = 11.84LQGSTLTQYY248 pKa = 10.43TFIQGQEE255 pKa = 3.91LAVKK259 pKa = 10.57FKK261 pKa = 11.03DD262 pKa = 3.33AA263 pKa = 4.38
MM1 pKa = 7.16YY2 pKa = 10.36GRR4 pKa = 11.84RR5 pKa = 11.84SYY7 pKa = 10.89RR8 pKa = 11.84RR9 pKa = 11.84YY10 pKa = 7.66RR11 pKa = 11.84TYY13 pKa = 10.96RR14 pKa = 11.84KK15 pKa = 8.91GSGRR19 pKa = 11.84ARR21 pKa = 11.84YY22 pKa = 8.39GATKK26 pKa = 9.64ARR28 pKa = 11.84RR29 pKa = 11.84VVRR32 pKa = 11.84YY33 pKa = 9.06RR34 pKa = 11.84GARR37 pKa = 11.84ARR39 pKa = 11.84GIKK42 pKa = 10.09RR43 pKa = 11.84IVNRR47 pKa = 11.84QIEE50 pKa = 4.61TKK52 pKa = 9.76EE53 pKa = 4.05YY54 pKa = 10.14TNGTQTSMLQPTQLGAFGAGYY75 pKa = 10.48NGFSDD80 pKa = 3.59TVSYY84 pKa = 8.6FKK86 pKa = 11.11PMDD89 pKa = 3.82YY90 pKa = 10.73LSQGTSRR97 pKa = 11.84NTRR100 pKa = 11.84IGDD103 pKa = 3.39AVTIMSIEE111 pKa = 3.53WRR113 pKa = 11.84YY114 pKa = 10.72CITFTDD120 pKa = 4.24YY121 pKa = 9.8EE122 pKa = 4.33QEE124 pKa = 4.58FIRR127 pKa = 11.84VMAFTAPFGFTPTYY141 pKa = 11.07GNIFEE146 pKa = 5.4APDD149 pKa = 3.51GNVDD153 pKa = 3.54VRR155 pKa = 11.84VARR158 pKa = 11.84PNTNITVLKK167 pKa = 9.91QYY169 pKa = 11.0KK170 pKa = 8.39HH171 pKa = 5.58TLSPLNSATASSSSTVNTPQVFRR194 pKa = 11.84TGCLRR199 pKa = 11.84LLFPGGRR206 pKa = 11.84KK207 pKa = 8.43FQYY210 pKa = 10.67SFNNYY215 pKa = 8.42TGSTALFNRR224 pKa = 11.84NRR226 pKa = 11.84DD227 pKa = 3.2IFIVAVPFAAKK238 pKa = 10.14RR239 pKa = 11.84LQGSTLTQYY248 pKa = 10.43TFIQGQEE255 pKa = 3.91LAVKK259 pKa = 10.57FKK261 pKa = 11.03DD262 pKa = 3.33AA263 pKa = 4.38
Molecular weight: 29.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
567 |
263 |
304 |
283.5 |
32.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.467 ± 1.329 | 1.235 ± 0.295 |
4.586 ± 0.96 | 4.762 ± 1.306 |
5.82 ± 0.637 | 7.407 ± 0.596 |
0.882 ± 0.312 | 3.704 ± 0.771 |
5.291 ± 0.689 | 5.996 ± 0.655 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.175 ± 0.792 | 5.467 ± 0.09 |
4.938 ± 0.706 | 4.938 ± 0.003 |
8.466 ± 1.356 | 5.82 ± 0.4 |
8.995 ± 1.027 | 6.173 ± 0.292 |
1.235 ± 0.531 | 5.644 ± 0.51 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
