
Capybara microvirus Cap3_SP_613
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.57
Get precalculated fractions of proteins
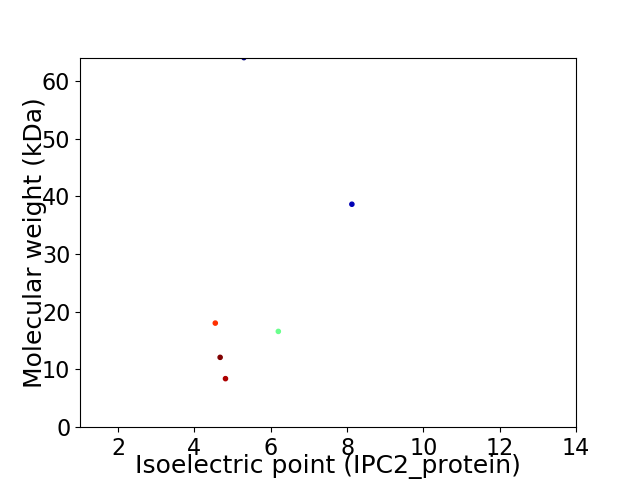
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
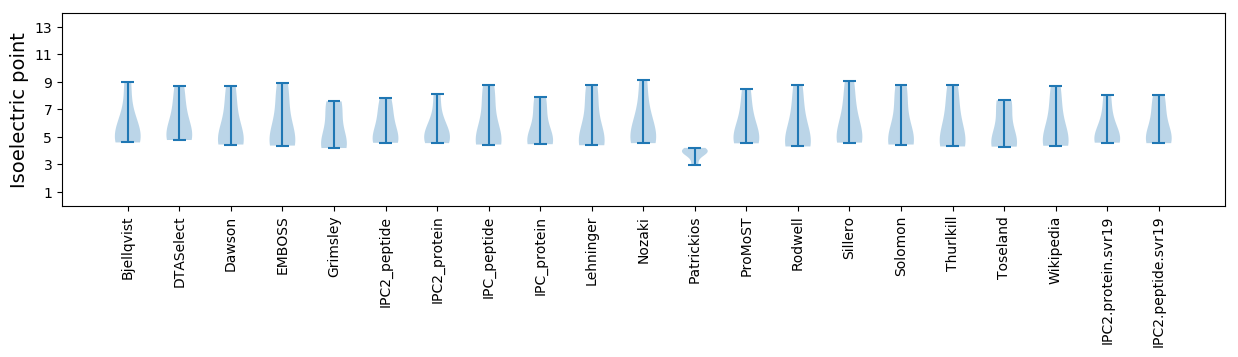
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W569|A0A4P8W569_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_613 OX=2585483 PE=3 SV=1
MM1 pKa = 7.58LKK3 pKa = 10.79LNLRR7 pKa = 11.84EE8 pKa = 3.88NFIYY12 pKa = 10.58EE13 pKa = 4.28SIDD16 pKa = 3.39YY17 pKa = 10.45SKK19 pKa = 11.51GSIVDD24 pKa = 3.68TYY26 pKa = 11.55KK27 pKa = 11.13VILPYY32 pKa = 10.68QKK34 pKa = 10.43DD35 pKa = 3.72DD36 pKa = 3.85KK37 pKa = 11.62GVFLNDD43 pKa = 3.3SDD45 pKa = 3.9TPIYY49 pKa = 10.42VKK51 pKa = 10.43SGKK54 pKa = 10.07KK55 pKa = 10.16NLDD58 pKa = 3.24EE59 pKa = 4.93YY60 pKa = 11.13INSFKK65 pKa = 10.93DD66 pKa = 2.82EE67 pKa = 4.11CDD69 pKa = 2.83IYY71 pKa = 11.33NILSKK76 pKa = 10.97FEE78 pKa = 4.13STGDD82 pKa = 3.56VSSLNLGRR90 pKa = 11.84GVYY93 pKa = 10.65ADD95 pKa = 3.35ISKK98 pKa = 10.57IPDD101 pKa = 3.97NIHH104 pKa = 7.23DD105 pKa = 4.55FNNALNQGAEE115 pKa = 4.13ILNTLDD121 pKa = 3.02SDD123 pKa = 3.72LRR125 pKa = 11.84KK126 pKa = 10.07AILEE130 pKa = 3.9NDD132 pKa = 2.79IQAINKK138 pKa = 6.94ATNNILNGSNKK149 pKa = 9.53EE150 pKa = 4.02VLQDD154 pKa = 3.55EE155 pKa = 4.97SIKK158 pKa = 10.65EE159 pKa = 3.91AA160 pKa = 3.86
MM1 pKa = 7.58LKK3 pKa = 10.79LNLRR7 pKa = 11.84EE8 pKa = 3.88NFIYY12 pKa = 10.58EE13 pKa = 4.28SIDD16 pKa = 3.39YY17 pKa = 10.45SKK19 pKa = 11.51GSIVDD24 pKa = 3.68TYY26 pKa = 11.55KK27 pKa = 11.13VILPYY32 pKa = 10.68QKK34 pKa = 10.43DD35 pKa = 3.72DD36 pKa = 3.85KK37 pKa = 11.62GVFLNDD43 pKa = 3.3SDD45 pKa = 3.9TPIYY49 pKa = 10.42VKK51 pKa = 10.43SGKK54 pKa = 10.07KK55 pKa = 10.16NLDD58 pKa = 3.24EE59 pKa = 4.93YY60 pKa = 11.13INSFKK65 pKa = 10.93DD66 pKa = 2.82EE67 pKa = 4.11CDD69 pKa = 2.83IYY71 pKa = 11.33NILSKK76 pKa = 10.97FEE78 pKa = 4.13STGDD82 pKa = 3.56VSSLNLGRR90 pKa = 11.84GVYY93 pKa = 10.65ADD95 pKa = 3.35ISKK98 pKa = 10.57IPDD101 pKa = 3.97NIHH104 pKa = 7.23DD105 pKa = 4.55FNNALNQGAEE115 pKa = 4.13ILNTLDD121 pKa = 3.02SDD123 pKa = 3.72LRR125 pKa = 11.84KK126 pKa = 10.07AILEE130 pKa = 3.9NDD132 pKa = 2.79IQAINKK138 pKa = 6.94ATNNILNGSNKK149 pKa = 9.53EE150 pKa = 4.02VLQDD154 pKa = 3.55EE155 pKa = 4.97SIKK158 pKa = 10.65EE159 pKa = 3.91AA160 pKa = 3.86
Molecular weight: 18.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W5S5|A0A4P8W5S5_9VIRU Minor capsid protein OS=Capybara microvirus Cap3_SP_613 OX=2585483 PE=4 SV=1
MM1 pKa = 7.31TCLNLSYY8 pKa = 10.69VPCIKK13 pKa = 10.63YY14 pKa = 10.58KK15 pKa = 10.99NSDD18 pKa = 3.33KK19 pKa = 11.32KK20 pKa = 11.47KK21 pKa = 8.8MLFKK25 pKa = 10.87KK26 pKa = 10.42KK27 pKa = 10.15ILVKK31 pKa = 10.72NKK33 pKa = 10.08SDD35 pKa = 4.44LDD37 pKa = 3.96LKK39 pKa = 10.46IDD41 pKa = 3.97KK42 pKa = 9.54EE43 pKa = 4.41NCFLEE48 pKa = 4.77LKK50 pKa = 10.18EE51 pKa = 4.27SSNLFDD57 pKa = 3.83SVEE60 pKa = 4.01FLQVPCGICKK70 pKa = 10.31EE71 pKa = 4.15CLKK74 pKa = 11.04KK75 pKa = 10.36NARR78 pKa = 11.84DD79 pKa = 3.12WAFRR83 pKa = 11.84CVKK86 pKa = 9.93EE87 pKa = 3.78ASQYY91 pKa = 10.2KK92 pKa = 8.09EE93 pKa = 3.83NYY95 pKa = 8.73MITLTYY101 pKa = 11.18DD102 pKa = 3.33NDD104 pKa = 3.61NLPFNGSLVKK114 pKa = 10.92DD115 pKa = 4.42EE116 pKa = 4.06ISKK119 pKa = 10.5FNKK122 pKa = 9.61KK123 pKa = 10.03LKK125 pKa = 9.77TYY127 pKa = 10.69LSRR130 pKa = 11.84LNLPSDD136 pKa = 3.6FRR138 pKa = 11.84FFGCGEE144 pKa = 4.01YY145 pKa = 10.69GSQFGRR151 pKa = 11.84PHH153 pKa = 4.51YY154 pKa = 10.34HH155 pKa = 6.47IIYY158 pKa = 10.56FNLQLSKK165 pKa = 10.9LSEE168 pKa = 4.25LLGLRR173 pKa = 11.84YY174 pKa = 9.75YY175 pKa = 10.88GKK177 pKa = 10.04SSKK180 pKa = 10.49GFPIYY185 pKa = 10.35SCKK188 pKa = 10.4FLDD191 pKa = 4.06DD192 pKa = 3.52TWSKK196 pKa = 11.38GIVKK200 pKa = 10.07VSEE203 pKa = 4.16CDD205 pKa = 3.03IGSCAYY211 pKa = 9.3VAHH214 pKa = 6.19YY215 pKa = 10.34VDD217 pKa = 4.73KK218 pKa = 10.87KK219 pKa = 10.49KK220 pKa = 10.98CFTKK224 pKa = 9.98GQKK227 pKa = 9.68EE228 pKa = 3.74FFYY231 pKa = 10.97NNTGLVPDD239 pKa = 4.36FCVMSRR245 pKa = 11.84NPGIGSCFIEE255 pKa = 5.06DD256 pKa = 3.32VDD258 pKa = 4.49NNFLKK263 pKa = 11.04GFFNAYY269 pKa = 10.72LNGQCYY275 pKa = 10.14AIPKK279 pKa = 9.75FYY281 pKa = 10.38INKK284 pKa = 9.73LKK286 pKa = 10.72DD287 pKa = 3.59YY288 pKa = 10.58GLTSYY293 pKa = 10.99DD294 pKa = 3.07IYY296 pKa = 11.33KK297 pKa = 10.99DD298 pKa = 3.45NLLSNRR304 pKa = 11.84FNIDD308 pKa = 3.05SYY310 pKa = 11.69FLDD313 pKa = 3.56IEE315 pKa = 4.57KK316 pKa = 10.78GFSLNYY322 pKa = 9.96NLSNNKK328 pKa = 8.62GRR330 pKa = 11.84CFF332 pKa = 3.45
MM1 pKa = 7.31TCLNLSYY8 pKa = 10.69VPCIKK13 pKa = 10.63YY14 pKa = 10.58KK15 pKa = 10.99NSDD18 pKa = 3.33KK19 pKa = 11.32KK20 pKa = 11.47KK21 pKa = 8.8MLFKK25 pKa = 10.87KK26 pKa = 10.42KK27 pKa = 10.15ILVKK31 pKa = 10.72NKK33 pKa = 10.08SDD35 pKa = 4.44LDD37 pKa = 3.96LKK39 pKa = 10.46IDD41 pKa = 3.97KK42 pKa = 9.54EE43 pKa = 4.41NCFLEE48 pKa = 4.77LKK50 pKa = 10.18EE51 pKa = 4.27SSNLFDD57 pKa = 3.83SVEE60 pKa = 4.01FLQVPCGICKK70 pKa = 10.31EE71 pKa = 4.15CLKK74 pKa = 11.04KK75 pKa = 10.36NARR78 pKa = 11.84DD79 pKa = 3.12WAFRR83 pKa = 11.84CVKK86 pKa = 9.93EE87 pKa = 3.78ASQYY91 pKa = 10.2KK92 pKa = 8.09EE93 pKa = 3.83NYY95 pKa = 8.73MITLTYY101 pKa = 11.18DD102 pKa = 3.33NDD104 pKa = 3.61NLPFNGSLVKK114 pKa = 10.92DD115 pKa = 4.42EE116 pKa = 4.06ISKK119 pKa = 10.5FNKK122 pKa = 9.61KK123 pKa = 10.03LKK125 pKa = 9.77TYY127 pKa = 10.69LSRR130 pKa = 11.84LNLPSDD136 pKa = 3.6FRR138 pKa = 11.84FFGCGEE144 pKa = 4.01YY145 pKa = 10.69GSQFGRR151 pKa = 11.84PHH153 pKa = 4.51YY154 pKa = 10.34HH155 pKa = 6.47IIYY158 pKa = 10.56FNLQLSKK165 pKa = 10.9LSEE168 pKa = 4.25LLGLRR173 pKa = 11.84YY174 pKa = 9.75YY175 pKa = 10.88GKK177 pKa = 10.04SSKK180 pKa = 10.49GFPIYY185 pKa = 10.35SCKK188 pKa = 10.4FLDD191 pKa = 4.06DD192 pKa = 3.52TWSKK196 pKa = 11.38GIVKK200 pKa = 10.07VSEE203 pKa = 4.16CDD205 pKa = 3.03IGSCAYY211 pKa = 9.3VAHH214 pKa = 6.19YY215 pKa = 10.34VDD217 pKa = 4.73KK218 pKa = 10.87KK219 pKa = 10.49KK220 pKa = 10.98CFTKK224 pKa = 9.98GQKK227 pKa = 9.68EE228 pKa = 3.74FFYY231 pKa = 10.97NNTGLVPDD239 pKa = 4.36FCVMSRR245 pKa = 11.84NPGIGSCFIEE255 pKa = 5.06DD256 pKa = 3.32VDD258 pKa = 4.49NNFLKK263 pKa = 11.04GFFNAYY269 pKa = 10.72LNGQCYY275 pKa = 10.14AIPKK279 pKa = 9.75FYY281 pKa = 10.38INKK284 pKa = 9.73LKK286 pKa = 10.72DD287 pKa = 3.59YY288 pKa = 10.58GLTSYY293 pKa = 10.99DD294 pKa = 3.07IYY296 pKa = 11.33KK297 pKa = 10.99DD298 pKa = 3.45NLLSNRR304 pKa = 11.84FNIDD308 pKa = 3.05SYY310 pKa = 11.69FLDD313 pKa = 3.56IEE315 pKa = 4.57KK316 pKa = 10.78GFSLNYY322 pKa = 9.96NLSNNKK328 pKa = 8.62GRR330 pKa = 11.84CFF332 pKa = 3.45
Molecular weight: 38.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1394 |
70 |
571 |
232.3 |
26.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.524 ± 1.622 | 1.865 ± 0.803 |
6.456 ± 0.942 | 4.304 ± 0.32 |
6.6 ± 0.923 | 5.452 ± 0.833 |
1.22 ± 0.44 | 6.6 ± 1.071 |
6.887 ± 1.848 | 9.469 ± 0.381 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.578 ± 0.321 | 9.684 ± 1.018 |
3.156 ± 0.973 | 3.156 ± 0.689 |
2.941 ± 0.393 | 9.828 ± 0.951 |
4.161 ± 0.686 | 4.878 ± 0.395 |
0.717 ± 0.257 | 5.524 ± 0.548 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
