
Vibrio virus Vf33
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Inoviridae; Villovirus
Average proteome isoelectric point is 5.88
Get precalculated fractions of proteins
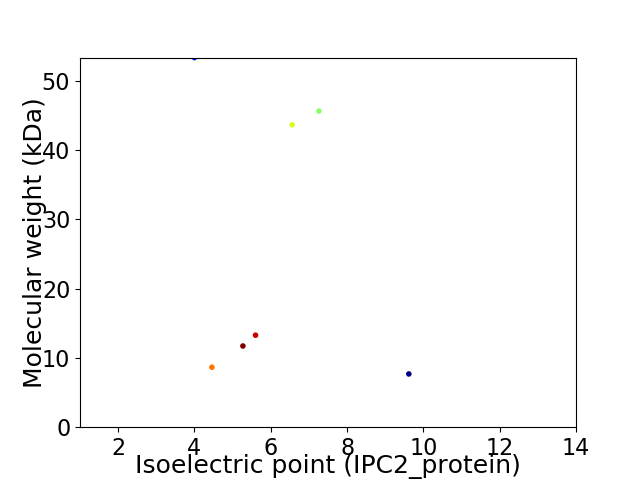
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q7DKP6|Q7DKP6_9VIRU Vpf77 OS=Vibrio virus Vf33 OX=127509 GN=vpf77 PE=4 SV=1
MM1 pKa = 6.89NHH3 pKa = 6.09YY4 pKa = 10.66LRR6 pKa = 11.84FFIALVILCASSHH19 pKa = 5.59TYY21 pKa = 10.64ALEE24 pKa = 3.73ARR26 pKa = 11.84IGYY29 pKa = 7.73MQMRR33 pKa = 11.84GCGSQGDD40 pKa = 3.71WVDD43 pKa = 3.54PYY45 pKa = 11.42KK46 pKa = 11.44VNTCFLDD53 pKa = 2.96TGYY56 pKa = 10.96FDD58 pKa = 4.08SCTFEE63 pKa = 3.84KK64 pKa = 10.36TPYY67 pKa = 10.25SDD69 pKa = 4.43ARR71 pKa = 11.84YY72 pKa = 8.79PYY74 pKa = 9.28QTVCDD79 pKa = 3.93NGLGLAYY86 pKa = 10.81YY87 pKa = 8.88EE88 pKa = 4.36VRR90 pKa = 11.84CPEE93 pKa = 3.81NSEE96 pKa = 4.27FDD98 pKa = 3.99PSTLRR103 pKa = 11.84CKK105 pKa = 10.33SVCEE109 pKa = 4.04YY110 pKa = 11.13GKK112 pKa = 10.26NPDD115 pKa = 3.94GTCMDD120 pKa = 3.88ACQFKK125 pKa = 10.9KK126 pKa = 10.85SIDD129 pKa = 3.7EE130 pKa = 4.31TKK132 pKa = 10.41LLQWVAYY139 pKa = 9.99VYY141 pKa = 10.92GEE143 pKa = 4.26QVTGACYY150 pKa = 10.82GDD152 pKa = 4.99FGATRR157 pKa = 11.84CEE159 pKa = 4.24LGRR162 pKa = 11.84VPSDD166 pKa = 3.07TTLCTDD172 pKa = 3.63VEE174 pKa = 4.7SGQWTQNTLCHH185 pKa = 6.58GNFQFTGNQCEE196 pKa = 4.28GGTLFWGKK204 pKa = 10.11DD205 pKa = 3.62GPDD208 pKa = 3.31TPIIPDD214 pKa = 4.37DD215 pKa = 5.14PIHH218 pKa = 7.61DD219 pKa = 4.88PDD221 pKa = 5.88DD222 pKa = 3.89PTGDD226 pKa = 3.49IEE228 pKa = 5.08DD229 pKa = 4.6PSVLPDD235 pKa = 3.61GSTNTVNPPDD245 pKa = 4.02TEE247 pKa = 4.11KK248 pKa = 11.1KK249 pKa = 10.17PDD251 pKa = 3.59VEE253 pKa = 6.26DD254 pKa = 5.31PDD256 pKa = 4.5TDD258 pKa = 4.12DD259 pKa = 3.73STDD262 pKa = 3.46MAVLNAIKK270 pKa = 10.53GLNSDD275 pKa = 3.77VNKK278 pKa = 10.59ALNDD282 pKa = 3.47MNIDD286 pKa = 3.46INQASADD293 pKa = 3.72VQNQIIALNASMVTNTQAIQKK314 pKa = 7.84QQINDD319 pKa = 3.14NKK321 pKa = 10.37IYY323 pKa = 10.49EE324 pKa = 4.16NTKK327 pKa = 10.95ALIQQANADD336 pKa = 3.12ITTAVNKK343 pKa = 7.74NTNAINGVGDD353 pKa = 4.15DD354 pKa = 3.78VEE356 pKa = 5.55KK357 pKa = 10.74IAGAMDD363 pKa = 5.45GIAEE367 pKa = 4.26DD368 pKa = 3.65VSGISDD374 pKa = 3.9TLDD377 pKa = 4.1GIANTDD383 pKa = 3.08TSGAGTGGTCIEE395 pKa = 4.51SQTCTGFYY403 pKa = 10.47EE404 pKa = 4.29SAYY407 pKa = 9.62PDD409 pKa = 3.66GLGGLVSGQLDD420 pKa = 3.78SLKK423 pKa = 11.03HH424 pKa = 4.14NTIDD428 pKa = 3.61NFVSSFGDD436 pKa = 3.42LDD438 pKa = 3.84LSSAKK443 pKa = 10.1RR444 pKa = 11.84PSFVLPVPFFGDD456 pKa = 3.76FSFEE460 pKa = 4.02EE461 pKa = 4.39QISFDD466 pKa = 3.32WVFGFIRR473 pKa = 11.84AVLIMTSVFAARR485 pKa = 11.84RR486 pKa = 11.84IIFGGG491 pKa = 3.39
MM1 pKa = 6.89NHH3 pKa = 6.09YY4 pKa = 10.66LRR6 pKa = 11.84FFIALVILCASSHH19 pKa = 5.59TYY21 pKa = 10.64ALEE24 pKa = 3.73ARR26 pKa = 11.84IGYY29 pKa = 7.73MQMRR33 pKa = 11.84GCGSQGDD40 pKa = 3.71WVDD43 pKa = 3.54PYY45 pKa = 11.42KK46 pKa = 11.44VNTCFLDD53 pKa = 2.96TGYY56 pKa = 10.96FDD58 pKa = 4.08SCTFEE63 pKa = 3.84KK64 pKa = 10.36TPYY67 pKa = 10.25SDD69 pKa = 4.43ARR71 pKa = 11.84YY72 pKa = 8.79PYY74 pKa = 9.28QTVCDD79 pKa = 3.93NGLGLAYY86 pKa = 10.81YY87 pKa = 8.88EE88 pKa = 4.36VRR90 pKa = 11.84CPEE93 pKa = 3.81NSEE96 pKa = 4.27FDD98 pKa = 3.99PSTLRR103 pKa = 11.84CKK105 pKa = 10.33SVCEE109 pKa = 4.04YY110 pKa = 11.13GKK112 pKa = 10.26NPDD115 pKa = 3.94GTCMDD120 pKa = 3.88ACQFKK125 pKa = 10.9KK126 pKa = 10.85SIDD129 pKa = 3.7EE130 pKa = 4.31TKK132 pKa = 10.41LLQWVAYY139 pKa = 9.99VYY141 pKa = 10.92GEE143 pKa = 4.26QVTGACYY150 pKa = 10.82GDD152 pKa = 4.99FGATRR157 pKa = 11.84CEE159 pKa = 4.24LGRR162 pKa = 11.84VPSDD166 pKa = 3.07TTLCTDD172 pKa = 3.63VEE174 pKa = 4.7SGQWTQNTLCHH185 pKa = 6.58GNFQFTGNQCEE196 pKa = 4.28GGTLFWGKK204 pKa = 10.11DD205 pKa = 3.62GPDD208 pKa = 3.31TPIIPDD214 pKa = 4.37DD215 pKa = 5.14PIHH218 pKa = 7.61DD219 pKa = 4.88PDD221 pKa = 5.88DD222 pKa = 3.89PTGDD226 pKa = 3.49IEE228 pKa = 5.08DD229 pKa = 4.6PSVLPDD235 pKa = 3.61GSTNTVNPPDD245 pKa = 4.02TEE247 pKa = 4.11KK248 pKa = 11.1KK249 pKa = 10.17PDD251 pKa = 3.59VEE253 pKa = 6.26DD254 pKa = 5.31PDD256 pKa = 4.5TDD258 pKa = 4.12DD259 pKa = 3.73STDD262 pKa = 3.46MAVLNAIKK270 pKa = 10.53GLNSDD275 pKa = 3.77VNKK278 pKa = 10.59ALNDD282 pKa = 3.47MNIDD286 pKa = 3.46INQASADD293 pKa = 3.72VQNQIIALNASMVTNTQAIQKK314 pKa = 7.84QQINDD319 pKa = 3.14NKK321 pKa = 10.37IYY323 pKa = 10.49EE324 pKa = 4.16NTKK327 pKa = 10.95ALIQQANADD336 pKa = 3.12ITTAVNKK343 pKa = 7.74NTNAINGVGDD353 pKa = 4.15DD354 pKa = 3.78VEE356 pKa = 5.55KK357 pKa = 10.74IAGAMDD363 pKa = 5.45GIAEE367 pKa = 4.26DD368 pKa = 3.65VSGISDD374 pKa = 3.9TLDD377 pKa = 4.1GIANTDD383 pKa = 3.08TSGAGTGGTCIEE395 pKa = 4.51SQTCTGFYY403 pKa = 10.47EE404 pKa = 4.29SAYY407 pKa = 9.62PDD409 pKa = 3.66GLGGLVSGQLDD420 pKa = 3.78SLKK423 pKa = 11.03HH424 pKa = 4.14NTIDD428 pKa = 3.61NFVSSFGDD436 pKa = 3.42LDD438 pKa = 3.84LSSAKK443 pKa = 10.1RR444 pKa = 11.84PSFVLPVPFFGDD456 pKa = 3.76FSFEE460 pKa = 4.02EE461 pKa = 4.39QISFDD466 pKa = 3.32WVFGFIRR473 pKa = 11.84AVLIMTSVFAARR485 pKa = 11.84RR486 pKa = 11.84IIFGGG491 pKa = 3.39
Molecular weight: 53.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q7DKP7|Q7DKP7_9VIRU Vpf81 OS=Vibrio virus Vf33 OX=127509 GN=vpf81 PE=4 SV=1
MM1 pKa = 7.39KK2 pKa = 10.4RR3 pKa = 11.84LNALKK8 pKa = 10.66KK9 pKa = 10.26FGKK12 pKa = 9.38QAAATVTVAVLSVPAMAAEE31 pKa = 5.12GGAADD36 pKa = 4.47PFSAIDD42 pKa = 3.74LSGVATKK49 pKa = 10.3IGAAGLVIVGITMAYY64 pKa = 9.95KK65 pKa = 10.65SITLAKK71 pKa = 9.96RR72 pKa = 11.84AVNKK76 pKa = 10.37AA77 pKa = 3.05
MM1 pKa = 7.39KK2 pKa = 10.4RR3 pKa = 11.84LNALKK8 pKa = 10.66KK9 pKa = 10.26FGKK12 pKa = 9.38QAAATVTVAVLSVPAMAAEE31 pKa = 5.12GGAADD36 pKa = 4.47PFSAIDD42 pKa = 3.74LSGVATKK49 pKa = 10.3IGAAGLVIVGITMAYY64 pKa = 9.95KK65 pKa = 10.65SITLAKK71 pKa = 9.96RR72 pKa = 11.84AVNKK76 pKa = 10.37AA77 pKa = 3.05
Molecular weight: 7.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1652 |
77 |
491 |
236.0 |
26.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.809 ± 1.576 | 2.119 ± 0.516 |
7.324 ± 1.021 | 4.661 ± 0.576 |
4.661 ± 0.303 | 7.203 ± 0.594 |
1.695 ± 0.535 | 6.235 ± 0.395 |
5.569 ± 0.849 | 8.232 ± 1.11 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.24 ± 0.613 | 4.116 ± 0.697 |
4.782 ± 0.288 | 3.753 ± 0.454 |
4.237 ± 0.864 | 5.932 ± 0.244 |
6.78 ± 0.493 | 6.901 ± 0.691 |
1.695 ± 0.332 | 4.056 ± 0.565 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
