
Tissierella praeacuta DSM 18095
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Tissierellia; Tissierellales; Tissierellaceae; Tissierella; Tissierella praeacuta
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
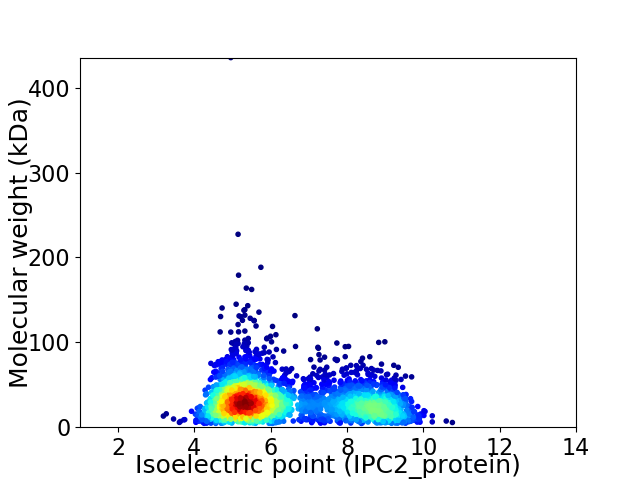
Virtual 2D-PAGE plot for 3095 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M4V032|A0A1M4V032_9FIRM Probable cell division protein WhiA OS=Tissierella praeacuta DSM 18095 OX=1123404 GN=whiA PE=3 SV=1
MM1 pKa = 7.73EE2 pKa = 5.93IIKK5 pKa = 10.31EE6 pKa = 4.06NSNSSIIIPIEE17 pKa = 3.78VLKK20 pKa = 11.08VSNLLDD26 pKa = 3.86SKK28 pKa = 11.16DD29 pKa = 3.66IEE31 pKa = 4.85LKK33 pKa = 9.03TLEE36 pKa = 4.11NAFVVMKK43 pKa = 9.81GTMNAMEE50 pKa = 5.4LIKK53 pKa = 10.62LAEE56 pKa = 4.17GLKK59 pKa = 10.41NLSEE63 pKa = 4.19EE64 pKa = 4.67LIEE67 pKa = 4.91HH68 pKa = 6.84LAGVCGRR75 pKa = 11.84CYY77 pKa = 10.6DD78 pKa = 3.39CSYY81 pKa = 11.1CEE83 pKa = 3.93RR84 pKa = 11.84FEE86 pKa = 4.54EE87 pKa = 4.23YY88 pKa = 10.91DD89 pKa = 3.79EE90 pKa = 4.98IVVPDD95 pKa = 4.0YY96 pKa = 11.18LLEE99 pKa = 4.24EE100 pKa = 4.37AGIPIDD106 pKa = 5.09AKK108 pKa = 9.95LCAYY112 pKa = 9.28TEE114 pKa = 4.14EE115 pKa = 4.84DD116 pKa = 3.15SGEE119 pKa = 4.28VVVIQADD126 pKa = 3.74YY127 pKa = 10.79DD128 pKa = 3.96YY129 pKa = 10.92DD130 pKa = 3.75IADD133 pKa = 3.55VPQFVIDD140 pKa = 3.39IFEE143 pKa = 4.06ISGICIRR150 pKa = 11.84EE151 pKa = 4.18LEE153 pKa = 4.41EE154 pKa = 3.74NLIMEE159 pKa = 4.94NIVYY163 pKa = 10.46GDD165 pKa = 3.53EE166 pKa = 4.04
MM1 pKa = 7.73EE2 pKa = 5.93IIKK5 pKa = 10.31EE6 pKa = 4.06NSNSSIIIPIEE17 pKa = 3.78VLKK20 pKa = 11.08VSNLLDD26 pKa = 3.86SKK28 pKa = 11.16DD29 pKa = 3.66IEE31 pKa = 4.85LKK33 pKa = 9.03TLEE36 pKa = 4.11NAFVVMKK43 pKa = 9.81GTMNAMEE50 pKa = 5.4LIKK53 pKa = 10.62LAEE56 pKa = 4.17GLKK59 pKa = 10.41NLSEE63 pKa = 4.19EE64 pKa = 4.67LIEE67 pKa = 4.91HH68 pKa = 6.84LAGVCGRR75 pKa = 11.84CYY77 pKa = 10.6DD78 pKa = 3.39CSYY81 pKa = 11.1CEE83 pKa = 3.93RR84 pKa = 11.84FEE86 pKa = 4.54EE87 pKa = 4.23YY88 pKa = 10.91DD89 pKa = 3.79EE90 pKa = 4.98IVVPDD95 pKa = 4.0YY96 pKa = 11.18LLEE99 pKa = 4.24EE100 pKa = 4.37AGIPIDD106 pKa = 5.09AKK108 pKa = 9.95LCAYY112 pKa = 9.28TEE114 pKa = 4.14EE115 pKa = 4.84DD116 pKa = 3.15SGEE119 pKa = 4.28VVVIQADD126 pKa = 3.74YY127 pKa = 10.79DD128 pKa = 3.96YY129 pKa = 10.92DD130 pKa = 3.75IADD133 pKa = 3.55VPQFVIDD140 pKa = 3.39IFEE143 pKa = 4.06ISGICIRR150 pKa = 11.84EE151 pKa = 4.18LEE153 pKa = 4.41EE154 pKa = 3.74NLIMEE159 pKa = 4.94NIVYY163 pKa = 10.46GDD165 pKa = 3.53EE166 pKa = 4.04
Molecular weight: 18.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M4SDJ3|A0A1M4SDJ3_9FIRM Geranylgeranyl diphosphate synthase type II OS=Tissierella praeacuta DSM 18095 OX=1123404 GN=SAMN02745784_00264 PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 10.0KK3 pKa = 10.47NKK5 pKa = 9.64IMTLGLSIALIIAMAGCTPKK25 pKa = 9.65GTNMRR30 pKa = 11.84NMSTQTRR37 pKa = 11.84LYY39 pKa = 10.66DD40 pKa = 3.53NNVNNRR46 pKa = 11.84WMTNAPFNARR56 pKa = 11.84DD57 pKa = 3.64RR58 pKa = 11.84LNTNLNNGMVRR69 pKa = 11.84NNSYY73 pKa = 11.32LNDD76 pKa = 3.45GVLRR80 pKa = 11.84DD81 pKa = 4.11NNYY84 pKa = 10.53LNNNVVRR91 pKa = 11.84DD92 pKa = 3.88NTLTNDD98 pKa = 3.13NIMRR102 pKa = 11.84NRR104 pKa = 11.84TNLDD108 pKa = 2.72SGMITNDD115 pKa = 2.4ISQLSTRR122 pKa = 11.84ADD124 pKa = 3.85AIARR128 pKa = 11.84RR129 pKa = 11.84VTALPEE135 pKa = 3.77VTGASVIVHH144 pKa = 6.67GNTAIVGCDD153 pKa = 3.15VRR155 pKa = 11.84GNTNNAISSNLKK167 pKa = 9.87QKK169 pKa = 10.72VEE171 pKa = 4.16AAVKK175 pKa = 8.81TADD178 pKa = 3.8RR179 pKa = 11.84NIKK182 pKa = 9.54KK183 pKa = 10.47VSVTSDD189 pKa = 3.03PSLYY193 pKa = 10.68SRR195 pKa = 11.84IRR197 pKa = 11.84TMSTSIGNGHH207 pKa = 7.62PISNFTRR214 pKa = 11.84DD215 pKa = 2.83IEE217 pKa = 5.04DD218 pKa = 2.96ILRR221 pKa = 11.84RR222 pKa = 11.84ITSPIRR228 pKa = 3.45
MM1 pKa = 7.67KK2 pKa = 10.0KK3 pKa = 10.47NKK5 pKa = 9.64IMTLGLSIALIIAMAGCTPKK25 pKa = 9.65GTNMRR30 pKa = 11.84NMSTQTRR37 pKa = 11.84LYY39 pKa = 10.66DD40 pKa = 3.53NNVNNRR46 pKa = 11.84WMTNAPFNARR56 pKa = 11.84DD57 pKa = 3.64RR58 pKa = 11.84LNTNLNNGMVRR69 pKa = 11.84NNSYY73 pKa = 11.32LNDD76 pKa = 3.45GVLRR80 pKa = 11.84DD81 pKa = 4.11NNYY84 pKa = 10.53LNNNVVRR91 pKa = 11.84DD92 pKa = 3.88NTLTNDD98 pKa = 3.13NIMRR102 pKa = 11.84NRR104 pKa = 11.84TNLDD108 pKa = 2.72SGMITNDD115 pKa = 2.4ISQLSTRR122 pKa = 11.84ADD124 pKa = 3.85AIARR128 pKa = 11.84RR129 pKa = 11.84VTALPEE135 pKa = 3.77VTGASVIVHH144 pKa = 6.67GNTAIVGCDD153 pKa = 3.15VRR155 pKa = 11.84GNTNNAISSNLKK167 pKa = 9.87QKK169 pKa = 10.72VEE171 pKa = 4.16AAVKK175 pKa = 8.81TADD178 pKa = 3.8RR179 pKa = 11.84NIKK182 pKa = 9.54KK183 pKa = 10.47VSVTSDD189 pKa = 3.03PSLYY193 pKa = 10.68SRR195 pKa = 11.84IRR197 pKa = 11.84TMSTSIGNGHH207 pKa = 7.62PISNFTRR214 pKa = 11.84DD215 pKa = 2.83IEE217 pKa = 5.04DD218 pKa = 2.96ILRR221 pKa = 11.84RR222 pKa = 11.84ITSPIRR228 pKa = 3.45
Molecular weight: 25.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
911760 |
39 |
3743 |
294.6 |
33.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.332 ± 0.057 | 0.891 ± 0.018 |
5.57 ± 0.038 | 7.834 ± 0.057 |
4.312 ± 0.036 | 6.675 ± 0.044 |
1.387 ± 0.017 | 10.669 ± 0.06 |
8.649 ± 0.046 | 9.25 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.71 ± 0.021 | 6.046 ± 0.043 |
2.935 ± 0.026 | 2.313 ± 0.024 |
3.662 ± 0.031 | 6.021 ± 0.033 |
4.877 ± 0.029 | 6.127 ± 0.036 |
0.679 ± 0.013 | 4.06 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
