
Capybara microvirus Cap3_SP_383
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.52
Get precalculated fractions of proteins
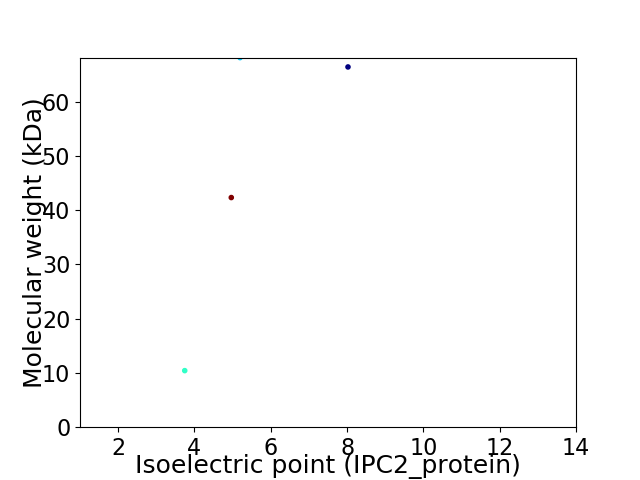
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W5R6|A0A4P8W5R6_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_383 OX=2585439 PE=4 SV=1
MM1 pKa = 7.59KK2 pKa = 10.56FNGFNGTDD10 pKa = 3.82IIQVPTPKK18 pKa = 10.13NPPMMLLQPNEE29 pKa = 4.14FGEE32 pKa = 4.38MEE34 pKa = 4.66LVAFDD39 pKa = 3.82DD40 pKa = 4.3TADD43 pKa = 3.55LGYY46 pKa = 11.12ASDD49 pKa = 4.07YY50 pKa = 11.28DD51 pKa = 3.58ITTLLEE57 pKa = 3.99NGIDD61 pKa = 3.56PRR63 pKa = 11.84FVGNTKK69 pKa = 8.97PTGSMLDD76 pKa = 3.26AYY78 pKa = 10.84SGLEE82 pKa = 3.83AFQPATNINEE92 pKa = 4.25TNEE95 pKa = 3.86
MM1 pKa = 7.59KK2 pKa = 10.56FNGFNGTDD10 pKa = 3.82IIQVPTPKK18 pKa = 10.13NPPMMLLQPNEE29 pKa = 4.14FGEE32 pKa = 4.38MEE34 pKa = 4.66LVAFDD39 pKa = 3.82DD40 pKa = 4.3TADD43 pKa = 3.55LGYY46 pKa = 11.12ASDD49 pKa = 4.07YY50 pKa = 11.28DD51 pKa = 3.58ITTLLEE57 pKa = 3.99NGIDD61 pKa = 3.56PRR63 pKa = 11.84FVGNTKK69 pKa = 8.97PTGSMLDD76 pKa = 3.26AYY78 pKa = 10.84SGLEE82 pKa = 3.83AFQPATNINEE92 pKa = 4.25TNEE95 pKa = 3.86
Molecular weight: 10.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W5Z5|A0A4P8W5Z5_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_383 OX=2585439 PE=3 SV=1
MM1 pKa = 8.21DD2 pKa = 5.38IFPKK6 pKa = 10.27SDD8 pKa = 2.56NHH10 pKa = 7.58ALIKK14 pKa = 10.41KK15 pKa = 9.65GYY17 pKa = 7.27TPNRR21 pKa = 11.84YY22 pKa = 9.7AMQIMNKK29 pKa = 7.25THH31 pKa = 6.27YY32 pKa = 9.43LHH34 pKa = 7.18PDD36 pKa = 2.33MRR38 pKa = 11.84YY39 pKa = 9.1RR40 pKa = 11.84YY41 pKa = 9.76NVINPYY47 pKa = 9.38SFCRR51 pKa = 11.84MFSQQQSYY59 pKa = 9.7YY60 pKa = 9.89IRR62 pKa = 11.84AYY64 pKa = 8.6YY65 pKa = 8.89QWLVTNEE72 pKa = 4.03QLGKK76 pKa = 9.28TIFLTFNYY84 pKa = 10.03NNNALPHH91 pKa = 6.08NYY93 pKa = 10.34GYY95 pKa = 7.84VTCDD99 pKa = 2.86NSHH102 pKa = 5.09IHH104 pKa = 6.52ALFRR108 pKa = 11.84NSYY111 pKa = 8.02LHH113 pKa = 7.05KK114 pKa = 10.37KK115 pKa = 9.16FKK117 pKa = 10.3DD118 pKa = 3.53YY119 pKa = 10.49RR120 pKa = 11.84IHH122 pKa = 7.36YY123 pKa = 8.34IICTEE128 pKa = 3.98FGEE131 pKa = 4.84GAGIRR136 pKa = 11.84KK137 pKa = 9.74KK138 pKa = 11.05GEE140 pKa = 3.8NPHH143 pKa = 5.06YY144 pKa = 10.28HH145 pKa = 6.11ALIYY149 pKa = 8.67LTHH152 pKa = 7.17RR153 pKa = 11.84NGEE156 pKa = 4.36CVTNHH161 pKa = 5.54VCRR164 pKa = 11.84QFLAEE169 pKa = 5.59CRR171 pKa = 11.84RR172 pKa = 11.84FWQGNPNPASKK183 pKa = 9.88TNDD186 pKa = 2.74PRR188 pKa = 11.84NFIFGKK194 pKa = 10.4VEE196 pKa = 3.81VAKK199 pKa = 11.04GKK201 pKa = 10.63GLFVDD206 pKa = 4.9SFHH209 pKa = 8.24SMQYY213 pKa = 8.88VSKK216 pKa = 9.55YY217 pKa = 8.01TIKK220 pKa = 11.04DD221 pKa = 2.82ADD223 pKa = 3.54TRR225 pKa = 11.84SRR227 pKa = 11.84MKK229 pKa = 10.48KK230 pKa = 9.75IEE232 pKa = 3.91DD233 pKa = 4.12AIYY236 pKa = 10.28NEE238 pKa = 3.24IRR240 pKa = 11.84TTVFRR245 pKa = 11.84DD246 pKa = 3.23EE247 pKa = 4.76SVNHH251 pKa = 5.65TLRR254 pKa = 11.84DD255 pKa = 3.56VLEE258 pKa = 3.99FHH260 pKa = 7.03PFVDD264 pKa = 4.75GVNDD268 pKa = 3.54EE269 pKa = 4.29QFNLSLPAHH278 pKa = 5.39YY279 pKa = 9.94FGYY282 pKa = 10.67EE283 pKa = 3.8LLHH286 pKa = 6.37YY287 pKa = 9.12YY288 pKa = 10.27QGVDD292 pKa = 2.74TGVRR296 pKa = 11.84GEE298 pKa = 4.18FKK300 pKa = 10.79SYY302 pKa = 9.3LTLLEE307 pKa = 4.27EE308 pKa = 4.2QVKK311 pKa = 10.03QLFEE315 pKa = 4.04EE316 pKa = 4.51RR317 pKa = 11.84LQEE320 pKa = 3.84EE321 pKa = 4.82KK322 pKa = 10.81KK323 pKa = 10.8RR324 pKa = 11.84IFPKK328 pKa = 10.44VFMSKK333 pKa = 10.36GFGSHH338 pKa = 6.16ALEE341 pKa = 5.82HH342 pKa = 6.18ITPDD346 pKa = 3.36YY347 pKa = 10.02KK348 pKa = 10.81IPISTPKK355 pKa = 10.32GIKK358 pKa = 9.45NVPLPMYY365 pKa = 10.29LYY367 pKa = 10.53RR368 pKa = 11.84KK369 pKa = 9.83LFYY372 pKa = 10.82DD373 pKa = 3.53VVKK376 pKa = 8.52TQEE379 pKa = 4.32GQVHH383 pKa = 4.74YY384 pKa = 9.93HH385 pKa = 5.26LNEE388 pKa = 4.05RR389 pKa = 11.84GVQYY393 pKa = 10.78KK394 pKa = 9.69IDD396 pKa = 3.81KK397 pKa = 9.94FDD399 pKa = 3.8NDD401 pKa = 3.96IYY403 pKa = 11.35NLRR406 pKa = 11.84QDD408 pKa = 3.4IYY410 pKa = 11.68AFVQNYY416 pKa = 10.0DD417 pKa = 4.3DD418 pKa = 4.41LRR420 pKa = 11.84EE421 pKa = 3.94MCDD424 pKa = 2.93IYY426 pKa = 11.21NKK428 pKa = 10.37NMYY431 pKa = 10.05DD432 pKa = 3.18SRR434 pKa = 11.84FINIYY439 pKa = 9.52HH440 pKa = 7.17AYY442 pKa = 10.02AVYY445 pKa = 10.15KK446 pKa = 9.86YY447 pKa = 9.53IYY449 pKa = 8.19EE450 pKa = 3.85GRR452 pKa = 11.84MFSLINPPQLDD463 pKa = 3.85FYY465 pKa = 11.2EE466 pKa = 4.33DD467 pKa = 3.69FKK469 pKa = 11.55RR470 pKa = 11.84FVLPHH475 pKa = 7.35DD476 pKa = 5.48DD477 pKa = 3.24EE478 pKa = 6.2DD479 pKa = 5.91LYY481 pKa = 12.05VNTLDD486 pKa = 4.75VDD488 pKa = 3.78EE489 pKa = 5.53AKK491 pKa = 10.75QNLIRR496 pKa = 11.84RR497 pKa = 11.84MRR499 pKa = 11.84LCDD502 pKa = 3.52YY503 pKa = 10.49AYY505 pKa = 10.57HH506 pKa = 7.21PDD508 pKa = 3.32FRR510 pKa = 11.84HH511 pKa = 5.08YY512 pKa = 10.65RR513 pKa = 11.84SLFEE517 pKa = 4.52TISDD521 pKa = 4.59RR522 pKa = 11.84IDD524 pKa = 3.4AYY526 pKa = 11.49NFDD529 pKa = 4.25KK530 pKa = 11.05DD531 pKa = 3.6AEE533 pKa = 4.25RR534 pKa = 11.84MKK536 pKa = 10.95SYY538 pKa = 10.74IEE540 pKa = 4.07RR541 pKa = 11.84KK542 pKa = 10.3NITTKK547 pKa = 10.44HH548 pKa = 5.84KK549 pKa = 9.6KK550 pKa = 7.56TLL552 pKa = 3.53
MM1 pKa = 8.21DD2 pKa = 5.38IFPKK6 pKa = 10.27SDD8 pKa = 2.56NHH10 pKa = 7.58ALIKK14 pKa = 10.41KK15 pKa = 9.65GYY17 pKa = 7.27TPNRR21 pKa = 11.84YY22 pKa = 9.7AMQIMNKK29 pKa = 7.25THH31 pKa = 6.27YY32 pKa = 9.43LHH34 pKa = 7.18PDD36 pKa = 2.33MRR38 pKa = 11.84YY39 pKa = 9.1RR40 pKa = 11.84YY41 pKa = 9.76NVINPYY47 pKa = 9.38SFCRR51 pKa = 11.84MFSQQQSYY59 pKa = 9.7YY60 pKa = 9.89IRR62 pKa = 11.84AYY64 pKa = 8.6YY65 pKa = 8.89QWLVTNEE72 pKa = 4.03QLGKK76 pKa = 9.28TIFLTFNYY84 pKa = 10.03NNNALPHH91 pKa = 6.08NYY93 pKa = 10.34GYY95 pKa = 7.84VTCDD99 pKa = 2.86NSHH102 pKa = 5.09IHH104 pKa = 6.52ALFRR108 pKa = 11.84NSYY111 pKa = 8.02LHH113 pKa = 7.05KK114 pKa = 10.37KK115 pKa = 9.16FKK117 pKa = 10.3DD118 pKa = 3.53YY119 pKa = 10.49RR120 pKa = 11.84IHH122 pKa = 7.36YY123 pKa = 8.34IICTEE128 pKa = 3.98FGEE131 pKa = 4.84GAGIRR136 pKa = 11.84KK137 pKa = 9.74KK138 pKa = 11.05GEE140 pKa = 3.8NPHH143 pKa = 5.06YY144 pKa = 10.28HH145 pKa = 6.11ALIYY149 pKa = 8.67LTHH152 pKa = 7.17RR153 pKa = 11.84NGEE156 pKa = 4.36CVTNHH161 pKa = 5.54VCRR164 pKa = 11.84QFLAEE169 pKa = 5.59CRR171 pKa = 11.84RR172 pKa = 11.84FWQGNPNPASKK183 pKa = 9.88TNDD186 pKa = 2.74PRR188 pKa = 11.84NFIFGKK194 pKa = 10.4VEE196 pKa = 3.81VAKK199 pKa = 11.04GKK201 pKa = 10.63GLFVDD206 pKa = 4.9SFHH209 pKa = 8.24SMQYY213 pKa = 8.88VSKK216 pKa = 9.55YY217 pKa = 8.01TIKK220 pKa = 11.04DD221 pKa = 2.82ADD223 pKa = 3.54TRR225 pKa = 11.84SRR227 pKa = 11.84MKK229 pKa = 10.48KK230 pKa = 9.75IEE232 pKa = 3.91DD233 pKa = 4.12AIYY236 pKa = 10.28NEE238 pKa = 3.24IRR240 pKa = 11.84TTVFRR245 pKa = 11.84DD246 pKa = 3.23EE247 pKa = 4.76SVNHH251 pKa = 5.65TLRR254 pKa = 11.84DD255 pKa = 3.56VLEE258 pKa = 3.99FHH260 pKa = 7.03PFVDD264 pKa = 4.75GVNDD268 pKa = 3.54EE269 pKa = 4.29QFNLSLPAHH278 pKa = 5.39YY279 pKa = 9.94FGYY282 pKa = 10.67EE283 pKa = 3.8LLHH286 pKa = 6.37YY287 pKa = 9.12YY288 pKa = 10.27QGVDD292 pKa = 2.74TGVRR296 pKa = 11.84GEE298 pKa = 4.18FKK300 pKa = 10.79SYY302 pKa = 9.3LTLLEE307 pKa = 4.27EE308 pKa = 4.2QVKK311 pKa = 10.03QLFEE315 pKa = 4.04EE316 pKa = 4.51RR317 pKa = 11.84LQEE320 pKa = 3.84EE321 pKa = 4.82KK322 pKa = 10.81KK323 pKa = 10.8RR324 pKa = 11.84IFPKK328 pKa = 10.44VFMSKK333 pKa = 10.36GFGSHH338 pKa = 6.16ALEE341 pKa = 5.82HH342 pKa = 6.18ITPDD346 pKa = 3.36YY347 pKa = 10.02KK348 pKa = 10.81IPISTPKK355 pKa = 10.32GIKK358 pKa = 9.45NVPLPMYY365 pKa = 10.29LYY367 pKa = 10.53RR368 pKa = 11.84KK369 pKa = 9.83LFYY372 pKa = 10.82DD373 pKa = 3.53VVKK376 pKa = 8.52TQEE379 pKa = 4.32GQVHH383 pKa = 4.74YY384 pKa = 9.93HH385 pKa = 5.26LNEE388 pKa = 4.05RR389 pKa = 11.84GVQYY393 pKa = 10.78KK394 pKa = 9.69IDD396 pKa = 3.81KK397 pKa = 9.94FDD399 pKa = 3.8NDD401 pKa = 3.96IYY403 pKa = 11.35NLRR406 pKa = 11.84QDD408 pKa = 3.4IYY410 pKa = 11.68AFVQNYY416 pKa = 10.0DD417 pKa = 4.3DD418 pKa = 4.41LRR420 pKa = 11.84EE421 pKa = 3.94MCDD424 pKa = 2.93IYY426 pKa = 11.21NKK428 pKa = 10.37NMYY431 pKa = 10.05DD432 pKa = 3.18SRR434 pKa = 11.84FINIYY439 pKa = 9.52HH440 pKa = 7.17AYY442 pKa = 10.02AVYY445 pKa = 10.15KK446 pKa = 9.86YY447 pKa = 9.53IYY449 pKa = 8.19EE450 pKa = 3.85GRR452 pKa = 11.84MFSLINPPQLDD463 pKa = 3.85FYY465 pKa = 11.2EE466 pKa = 4.33DD467 pKa = 3.69FKK469 pKa = 11.55RR470 pKa = 11.84FVLPHH475 pKa = 7.35DD476 pKa = 5.48DD477 pKa = 3.24EE478 pKa = 6.2DD479 pKa = 5.91LYY481 pKa = 12.05VNTLDD486 pKa = 4.75VDD488 pKa = 3.78EE489 pKa = 5.53AKK491 pKa = 10.75QNLIRR496 pKa = 11.84RR497 pKa = 11.84MRR499 pKa = 11.84LCDD502 pKa = 3.52YY503 pKa = 10.49AYY505 pKa = 10.57HH506 pKa = 7.21PDD508 pKa = 3.32FRR510 pKa = 11.84HH511 pKa = 5.08YY512 pKa = 10.65RR513 pKa = 11.84SLFEE517 pKa = 4.52TISDD521 pKa = 4.59RR522 pKa = 11.84IDD524 pKa = 3.4AYY526 pKa = 11.49NFDD529 pKa = 4.25KK530 pKa = 11.05DD531 pKa = 3.6AEE533 pKa = 4.25RR534 pKa = 11.84MKK536 pKa = 10.95SYY538 pKa = 10.74IEE540 pKa = 4.07RR541 pKa = 11.84KK542 pKa = 10.3NITTKK547 pKa = 10.44HH548 pKa = 5.84KK549 pKa = 9.6KK550 pKa = 7.56TLL552 pKa = 3.53
Molecular weight: 66.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1629 |
95 |
602 |
407.3 |
46.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.262 ± 1.376 | 0.798 ± 0.276 |
7.612 ± 0.626 | 4.911 ± 1.162 |
5.279 ± 0.86 | 5.893 ± 0.795 |
2.21 ± 1.107 | 5.402 ± 0.493 |
6.016 ± 0.687 | 8.165 ± 0.619 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.762 ± 0.407 | 7.428 ± 1.315 |
3.376 ± 1.055 | 4.604 ± 1.054 |
5.034 ± 0.81 | 5.955 ± 1.114 |
6.446 ± 0.996 | 4.85 ± 0.587 |
0.859 ± 0.292 | 6.139 ± 1.493 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
