
Pararobbsia alpina
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiaceae; Pararobbsia
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
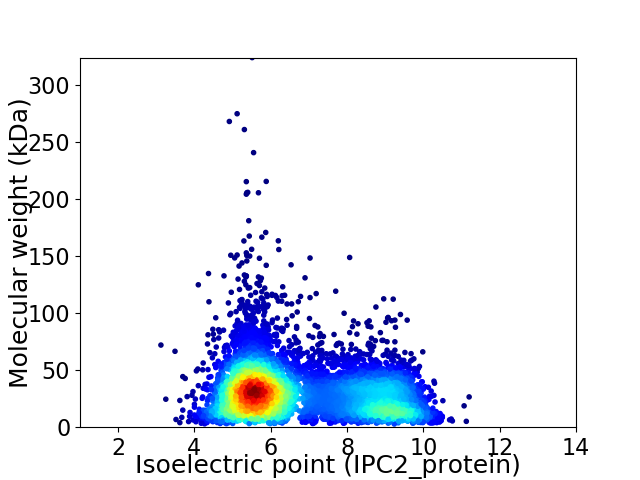
Virtual 2D-PAGE plot for 6080 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6S7B4P0|A0A6S7B4P0_9BURK Rieske domain-containing protein OS=Pararobbsia alpina OX=621374 GN=LMG28138_02334 PE=4 SV=1
MM1 pKa = 6.68KK2 pKa = 10.06TFWVWLLSAVLLGSVAACGGGHH24 pKa = 7.47DD25 pKa = 4.45DD26 pKa = 5.43HH27 pKa = 6.66STQRR31 pKa = 11.84PFGVAGIMEE40 pKa = 5.33GIWNGNLHH48 pKa = 6.59SNLNGGDD55 pKa = 2.79TGMFVIVTADD65 pKa = 3.36GALSLITNDD74 pKa = 3.8CQQISANIAANGPFFSGAGTTYY96 pKa = 10.87LQTNCDD102 pKa = 3.65GTDD105 pKa = 3.3VTLVSPTASAGPVQPFQIVGQFDD128 pKa = 4.29DD129 pKa = 3.69RR130 pKa = 11.84TGTAFANYY138 pKa = 6.82TTADD142 pKa = 3.59DD143 pKa = 3.89SGTISFLNFYY153 pKa = 9.71PDD155 pKa = 3.52YY156 pKa = 10.52FNEE159 pKa = 4.7PGTLPRR165 pKa = 11.84AAGAYY170 pKa = 10.31SFTQTLTALSIDD182 pKa = 3.64VNGILSYY189 pKa = 11.02RR190 pKa = 11.84DD191 pKa = 3.14AAGQAFPGTLFVIDD205 pKa = 4.39PTVDD209 pKa = 3.02AYY211 pKa = 10.96GMTLQVGSQTLTGLATLVDD230 pKa = 4.65DD231 pKa = 5.72GSGRR235 pKa = 11.84DD236 pKa = 3.45NDD238 pKa = 3.87FLFAVADD245 pKa = 4.05GNLAYY250 pKa = 10.57SAVLKK255 pKa = 10.66RR256 pKa = 11.84DD257 pKa = 3.15
MM1 pKa = 6.68KK2 pKa = 10.06TFWVWLLSAVLLGSVAACGGGHH24 pKa = 7.47DD25 pKa = 4.45DD26 pKa = 5.43HH27 pKa = 6.66STQRR31 pKa = 11.84PFGVAGIMEE40 pKa = 5.33GIWNGNLHH48 pKa = 6.59SNLNGGDD55 pKa = 2.79TGMFVIVTADD65 pKa = 3.36GALSLITNDD74 pKa = 3.8CQQISANIAANGPFFSGAGTTYY96 pKa = 10.87LQTNCDD102 pKa = 3.65GTDD105 pKa = 3.3VTLVSPTASAGPVQPFQIVGQFDD128 pKa = 4.29DD129 pKa = 3.69RR130 pKa = 11.84TGTAFANYY138 pKa = 6.82TTADD142 pKa = 3.59DD143 pKa = 3.89SGTISFLNFYY153 pKa = 9.71PDD155 pKa = 3.52YY156 pKa = 10.52FNEE159 pKa = 4.7PGTLPRR165 pKa = 11.84AAGAYY170 pKa = 10.31SFTQTLTALSIDD182 pKa = 3.64VNGILSYY189 pKa = 11.02RR190 pKa = 11.84DD191 pKa = 3.14AAGQAFPGTLFVIDD205 pKa = 4.39PTVDD209 pKa = 3.02AYY211 pKa = 10.96GMTLQVGSQTLTGLATLVDD230 pKa = 4.65DD231 pKa = 5.72GSGRR235 pKa = 11.84DD236 pKa = 3.45NDD238 pKa = 3.87FLFAVADD245 pKa = 4.05GNLAYY250 pKa = 10.57SAVLKK255 pKa = 10.66RR256 pKa = 11.84DD257 pKa = 3.15
Molecular weight: 26.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6S7CUD4|A0A6S7CUD4_9BURK Lipopolysaccharide export system protein LptA OS=Pararobbsia alpina OX=621374 GN=lptA PE=3 SV=1
MM1 pKa = 7.48RR2 pKa = 11.84AAHH5 pKa = 6.59RR6 pKa = 11.84FARR9 pKa = 11.84VHH11 pKa = 6.09RR12 pKa = 11.84ASPCRR17 pKa = 11.84TDD19 pKa = 3.24RR20 pKa = 11.84RR21 pKa = 11.84QAMRR25 pKa = 11.84RR26 pKa = 11.84APSVRR31 pKa = 11.84TLARR35 pKa = 11.84AAHH38 pKa = 6.48ANRR41 pKa = 11.84GPMAMRR47 pKa = 11.84HH48 pKa = 4.28VHH50 pKa = 6.46LALTSTHH57 pKa = 6.3RR58 pKa = 11.84ASRR61 pKa = 11.84APMVTRR67 pKa = 11.84HH68 pKa = 5.04VHH70 pKa = 5.38PVPTSMVRR78 pKa = 11.84ASRR81 pKa = 11.84APMVTRR87 pKa = 11.84HH88 pKa = 5.04VHH90 pKa = 5.38PVPTSMVRR98 pKa = 11.84ASRR101 pKa = 11.84APMVTRR107 pKa = 11.84HH108 pKa = 5.85VYY110 pKa = 9.21PVPTSMVRR118 pKa = 11.84ASRR121 pKa = 11.84APMVTRR127 pKa = 11.84RR128 pKa = 11.84VHH130 pKa = 5.9PVPTSMARR138 pKa = 11.84ASRR141 pKa = 11.84APMVTRR147 pKa = 11.84RR148 pKa = 11.84AHH150 pKa = 5.79PVPTSMARR158 pKa = 11.84ASRR161 pKa = 11.84APMVTPRR168 pKa = 11.84VHH170 pKa = 6.73PVPTSMARR178 pKa = 11.84ASRR181 pKa = 11.84APMVTRR187 pKa = 11.84HH188 pKa = 5.39VRR190 pKa = 11.84LAPTSMVRR198 pKa = 11.84VSRR201 pKa = 11.84VPMVMSLVHH210 pKa = 6.39LVAISIPRR218 pKa = 11.84AGRR221 pKa = 11.84VRR223 pKa = 11.84VAMRR227 pKa = 11.84GVPQAMRR234 pKa = 11.84HH235 pKa = 4.86LDD237 pKa = 3.33ARR239 pKa = 4.21
MM1 pKa = 7.48RR2 pKa = 11.84AAHH5 pKa = 6.59RR6 pKa = 11.84FARR9 pKa = 11.84VHH11 pKa = 6.09RR12 pKa = 11.84ASPCRR17 pKa = 11.84TDD19 pKa = 3.24RR20 pKa = 11.84RR21 pKa = 11.84QAMRR25 pKa = 11.84RR26 pKa = 11.84APSVRR31 pKa = 11.84TLARR35 pKa = 11.84AAHH38 pKa = 6.48ANRR41 pKa = 11.84GPMAMRR47 pKa = 11.84HH48 pKa = 4.28VHH50 pKa = 6.46LALTSTHH57 pKa = 6.3RR58 pKa = 11.84ASRR61 pKa = 11.84APMVTRR67 pKa = 11.84HH68 pKa = 5.04VHH70 pKa = 5.38PVPTSMVRR78 pKa = 11.84ASRR81 pKa = 11.84APMVTRR87 pKa = 11.84HH88 pKa = 5.04VHH90 pKa = 5.38PVPTSMVRR98 pKa = 11.84ASRR101 pKa = 11.84APMVTRR107 pKa = 11.84HH108 pKa = 5.85VYY110 pKa = 9.21PVPTSMVRR118 pKa = 11.84ASRR121 pKa = 11.84APMVTRR127 pKa = 11.84RR128 pKa = 11.84VHH130 pKa = 5.9PVPTSMARR138 pKa = 11.84ASRR141 pKa = 11.84APMVTRR147 pKa = 11.84RR148 pKa = 11.84AHH150 pKa = 5.79PVPTSMARR158 pKa = 11.84ASRR161 pKa = 11.84APMVTPRR168 pKa = 11.84VHH170 pKa = 6.73PVPTSMARR178 pKa = 11.84ASRR181 pKa = 11.84APMVTRR187 pKa = 11.84HH188 pKa = 5.39VRR190 pKa = 11.84LAPTSMVRR198 pKa = 11.84VSRR201 pKa = 11.84VPMVMSLVHH210 pKa = 6.39LVAISIPRR218 pKa = 11.84AGRR221 pKa = 11.84VRR223 pKa = 11.84VAMRR227 pKa = 11.84GVPQAMRR234 pKa = 11.84HH235 pKa = 4.86LDD237 pKa = 3.33ARR239 pKa = 4.21
Molecular weight: 26.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1865431 |
29 |
2929 |
306.8 |
33.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.191 ± 0.038 | 0.927 ± 0.009 |
5.448 ± 0.022 | 5.235 ± 0.03 |
3.668 ± 0.02 | 8.186 ± 0.029 |
2.353 ± 0.016 | 4.891 ± 0.023 |
3.106 ± 0.028 | 10.264 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.38 ± 0.014 | 2.824 ± 0.021 |
5.085 ± 0.024 | 3.594 ± 0.022 |
6.959 ± 0.031 | 6.034 ± 0.024 |
5.481 ± 0.026 | 7.666 ± 0.026 |
1.344 ± 0.012 | 2.365 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
