
Kineococcus radiotolerans (strain ATCC BAA-149 / DSM 14245 / SRS30216)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Kineosporiales; Kineosporiaceae; Kineococcus; Kineococcus radiotolerans
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
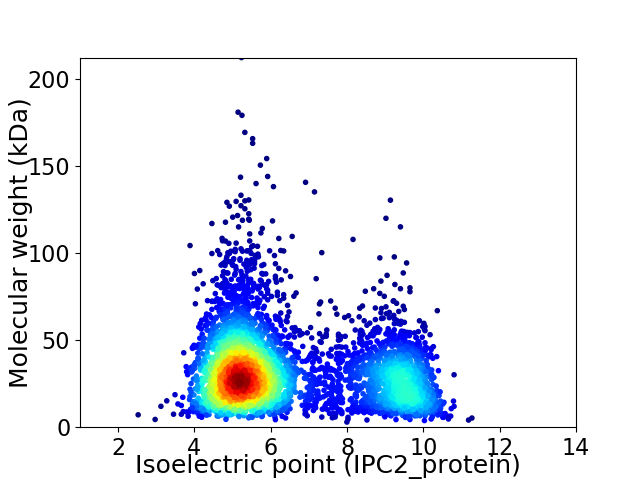
Virtual 2D-PAGE plot for 4674 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
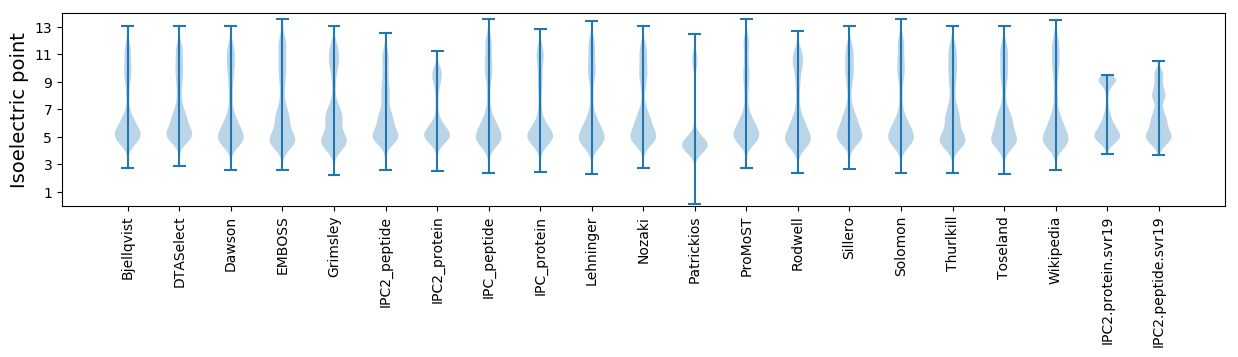
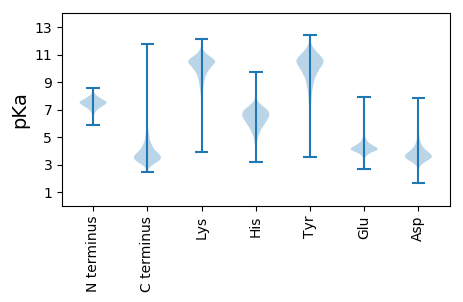
Protein with the lowest isoelectric point:
>tr|A6WF53|A6WF53_KINRD Exodeoxyribonuclease III OS=Kineococcus radiotolerans (strain ATCC BAA-149 / DSM 14245 / SRS30216) OX=266940 GN=Krad_3979 PE=3 SV=1
MM1 pKa = 7.18TNHH4 pKa = 6.39RR5 pKa = 11.84PSYY8 pKa = 9.76EE9 pKa = 3.83GRR11 pKa = 11.84PLPRR15 pKa = 11.84PAEE18 pKa = 4.32DD19 pKa = 4.36VDD21 pKa = 4.04DD22 pKa = 4.07QGLAFDD28 pKa = 5.15LATVATRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84ALGLLGLGAAGLTLAACGDD56 pKa = 4.12DD57 pKa = 3.48ATASSSTSSSAPTSGALTEE76 pKa = 4.91IPDD79 pKa = 3.97EE80 pKa = 4.36TAGPYY85 pKa = 10.2PGDD88 pKa = 3.58GSNGVDD94 pKa = 3.16VLEE97 pKa = 4.47RR98 pKa = 11.84SGVVRR103 pKa = 11.84SDD105 pKa = 2.68IRR107 pKa = 11.84SSFGTSTTTAEE118 pKa = 4.42GVPMTLTLTVVDD130 pKa = 4.43LAGGNVPFEE139 pKa = 4.26GVAVYY144 pKa = 9.93VWHH147 pKa = 7.6CDD149 pKa = 3.18RR150 pKa = 11.84DD151 pKa = 4.18GEE153 pKa = 4.27YY154 pKa = 10.88SLYY157 pKa = 10.93SEE159 pKa = 6.18DD160 pKa = 4.83IADD163 pKa = 3.7EE164 pKa = 4.34NYY166 pKa = 10.64LRR168 pKa = 11.84GVQVADD174 pKa = 3.4ADD176 pKa = 4.32GNVTFTSVFPACYY189 pKa = 9.56DD190 pKa = 3.41GRR192 pKa = 11.84WPHH195 pKa = 5.21VHH197 pKa = 6.5FEE199 pKa = 4.27VYY201 pKa = 10.21PDD203 pKa = 3.78AEE205 pKa = 4.84SITDD209 pKa = 3.55VANCIATSQVALPQEE224 pKa = 4.22ACEE227 pKa = 4.05AVYY230 pKa = 8.64ATTGYY235 pKa = 8.88EE236 pKa = 3.81QSVTNLARR244 pKa = 11.84VTLATDD250 pKa = 3.33NVFGDD255 pKa = 4.76DD256 pKa = 4.09GGASQTPTSSGDD268 pKa = 3.25VTNGYY273 pKa = 10.59AIALTVGVDD282 pKa = 3.39TTTEE286 pKa = 3.98PTAGTMAGMGSDD298 pKa = 3.54SSGHH302 pKa = 6.45PGGGPGGGGTPPSGAPAAAVSAAA325 pKa = 3.57
MM1 pKa = 7.18TNHH4 pKa = 6.39RR5 pKa = 11.84PSYY8 pKa = 9.76EE9 pKa = 3.83GRR11 pKa = 11.84PLPRR15 pKa = 11.84PAEE18 pKa = 4.32DD19 pKa = 4.36VDD21 pKa = 4.04DD22 pKa = 4.07QGLAFDD28 pKa = 5.15LATVATRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84ALGLLGLGAAGLTLAACGDD56 pKa = 4.12DD57 pKa = 3.48ATASSSTSSSAPTSGALTEE76 pKa = 4.91IPDD79 pKa = 3.97EE80 pKa = 4.36TAGPYY85 pKa = 10.2PGDD88 pKa = 3.58GSNGVDD94 pKa = 3.16VLEE97 pKa = 4.47RR98 pKa = 11.84SGVVRR103 pKa = 11.84SDD105 pKa = 2.68IRR107 pKa = 11.84SSFGTSTTTAEE118 pKa = 4.42GVPMTLTLTVVDD130 pKa = 4.43LAGGNVPFEE139 pKa = 4.26GVAVYY144 pKa = 9.93VWHH147 pKa = 7.6CDD149 pKa = 3.18RR150 pKa = 11.84DD151 pKa = 4.18GEE153 pKa = 4.27YY154 pKa = 10.88SLYY157 pKa = 10.93SEE159 pKa = 6.18DD160 pKa = 4.83IADD163 pKa = 3.7EE164 pKa = 4.34NYY166 pKa = 10.64LRR168 pKa = 11.84GVQVADD174 pKa = 3.4ADD176 pKa = 4.32GNVTFTSVFPACYY189 pKa = 9.56DD190 pKa = 3.41GRR192 pKa = 11.84WPHH195 pKa = 5.21VHH197 pKa = 6.5FEE199 pKa = 4.27VYY201 pKa = 10.21PDD203 pKa = 3.78AEE205 pKa = 4.84SITDD209 pKa = 3.55VANCIATSQVALPQEE224 pKa = 4.22ACEE227 pKa = 4.05AVYY230 pKa = 8.64ATTGYY235 pKa = 8.88EE236 pKa = 3.81QSVTNLARR244 pKa = 11.84VTLATDD250 pKa = 3.33NVFGDD255 pKa = 4.76DD256 pKa = 4.09GGASQTPTSSGDD268 pKa = 3.25VTNGYY273 pKa = 10.59AIALTVGVDD282 pKa = 3.39TTTEE286 pKa = 3.98PTAGTMAGMGSDD298 pKa = 3.54SSGHH302 pKa = 6.45PGGGPGGGGTPPSGAPAAAVSAAA325 pKa = 3.57
Molecular weight: 32.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A6WGN4|A6WGN4_KINRD Uncharacterized protein OS=Kineococcus radiotolerans (strain ATCC BAA-149 / DSM 14245 / SRS30216) OX=266940 GN=Krad_4514 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.45VHH17 pKa = 5.31GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSARR35 pKa = 11.84RR36 pKa = 11.84TKK38 pKa = 10.67GRR40 pKa = 11.84AQISAA45 pKa = 3.65
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.45VHH17 pKa = 5.31GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSARR35 pKa = 11.84RR36 pKa = 11.84TKK38 pKa = 10.67GRR40 pKa = 11.84AQISAA45 pKa = 3.65
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1488609 |
27 |
1995 |
318.5 |
33.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.636 ± 0.044 | 0.645 ± 0.009 |
6.031 ± 0.029 | 5.374 ± 0.036 |
2.543 ± 0.019 | 9.545 ± 0.034 |
2.221 ± 0.019 | 2.397 ± 0.026 |
1.336 ± 0.02 | 10.754 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.367 ± 0.012 | 1.436 ± 0.017 |
6.17 ± 0.032 | 2.736 ± 0.022 |
8.447 ± 0.039 | 5.086 ± 0.022 |
6.167 ± 0.028 | 10.046 ± 0.04 |
1.469 ± 0.014 | 1.594 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
