
Pelagibacter phage HTVC201P
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Pelagivirus; unclassified Pelagivirus
Average proteome isoelectric point is 7.15
Get precalculated fractions of proteins
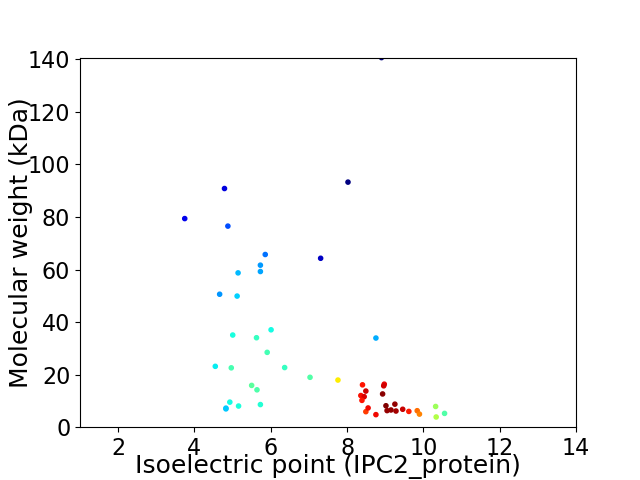
Virtual 2D-PAGE plot for 52 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y1NU81|A0A4Y1NU81_9CAUD Uncharacterized protein OS=Pelagibacter phage HTVC201P OX=2283024 GN=P201_gp50 PE=4 SV=1
MM1 pKa = 7.47ANSFVRR7 pKa = 11.84YY8 pKa = 7.53TGNNSTTSYY17 pKa = 10.59SIPFSYY23 pKa = 10.5RR24 pKa = 11.84ATSDD28 pKa = 3.24LTVTLSGVVTTAYY41 pKa = 9.21TLNSAGTTLTFNTAPAQDD59 pKa = 3.47VAIEE63 pKa = 3.8IRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84TSQGTKK73 pKa = 10.07LVDD76 pKa = 3.56YY77 pKa = 10.83ASGSVLTEE85 pKa = 3.68NDD87 pKa = 3.94LDD89 pKa = 4.06TDD91 pKa = 3.63SDD93 pKa = 3.8QAFFMSQEE101 pKa = 4.56AIDD104 pKa = 4.32DD105 pKa = 4.22AGDD108 pKa = 3.66VIKK111 pKa = 11.09VSNTNFQWDD120 pKa = 3.95VQNKK124 pKa = 9.66RR125 pKa = 11.84LTNVANPVNDD135 pKa = 3.36TDD137 pKa = 4.1AVNKK141 pKa = 9.93QFISTNLPNITTVAGISADD160 pKa = 3.48VTTVAGISSNVTTVANDD177 pKa = 3.35TTNIGTVATNMPSITTVATNINDD200 pKa = 3.93VIKK203 pKa = 11.06VADD206 pKa = 4.73DD207 pKa = 3.71LNEE210 pKa = 4.8AISEE214 pKa = 4.4VEE216 pKa = 4.24TVANDD221 pKa = 3.58LNEE224 pKa = 4.22ATSEE228 pKa = 4.16IEE230 pKa = 4.1VVANNIVNVNTVGTINADD248 pKa = 3.27VTTVANNEE256 pKa = 3.83TDD258 pKa = 3.26IQTLADD264 pKa = 4.24LEE266 pKa = 5.04DD267 pKa = 4.01GTVTTNGLSTLAGLNTEE284 pKa = 3.89IQGVYY289 pKa = 10.52NIRR292 pKa = 11.84TNVTNVDD299 pKa = 3.69TNSANVNLVAGQISPTNNVSAVSAVATEE327 pKa = 3.92IGTLGALGTEE337 pKa = 3.91ITNLNNIRR345 pKa = 11.84TDD347 pKa = 2.9ITGVNNIAPAVTAVNNNSANINTVAGLDD375 pKa = 3.74TEE377 pKa = 4.45ITALGASGTVASINTVASNLASVNNFGEE405 pKa = 4.61VYY407 pKa = 10.0RR408 pKa = 11.84ISATAPTTSLDD419 pKa = 3.8NGDD422 pKa = 3.17MWFDD426 pKa = 3.44TTAGKK431 pKa = 10.42LKK433 pKa = 9.75IWNGSSFDD441 pKa = 4.06LAGSSINGTSARR453 pKa = 11.84FKK455 pKa = 9.34YY456 pKa = 8.74TATASQTTFTGADD469 pKa = 3.49DD470 pKa = 4.13NGNTLAYY477 pKa = 10.26DD478 pKa = 3.44SGFTDD483 pKa = 3.98VYY485 pKa = 11.02LNGVKK490 pKa = 10.07LVNGSDD496 pKa = 3.53YY497 pKa = 10.98TATDD501 pKa = 3.61GSNVVLTTGASVNDD515 pKa = 3.34ILEE518 pKa = 4.12IVGFGTFSVADD529 pKa = 4.37FDD531 pKa = 5.44ASGLTGTINIARR543 pKa = 11.84LADD546 pKa = 3.58NSVTNAKK553 pKa = 9.64LANQSITINGSAVNLGGSVTVGEE576 pKa = 4.52TKK578 pKa = 9.18PTISSISPSTITNAQTSITITGSNFTSVPQVEE610 pKa = 4.76FLNPSTGIWYY620 pKa = 9.82VADD623 pKa = 3.22TVTYY627 pKa = 10.71NNSTSLTVNATLGVDD642 pKa = 3.59GTYY645 pKa = 10.3KK646 pKa = 10.15IRR648 pKa = 11.84IEE650 pKa = 4.26NPDD653 pKa = 3.26GNAVISSTNILTVSDD668 pKa = 3.76APTWTTASGTLGTFAGDD685 pKa = 3.63FSGTLATVVATSDD698 pKa = 3.47SAVTFSEE705 pKa = 4.29VGSNLATANVTLSSGGVLSTTDD727 pKa = 3.79FGGSSTTATTYY738 pKa = 11.13NFTIRR743 pKa = 11.84ATDD746 pKa = 3.77AEE748 pKa = 4.44NQTADD753 pKa = 3.8RR754 pKa = 11.84NFSLTSSFGATGGGQFNN771 pKa = 4.04
MM1 pKa = 7.47ANSFVRR7 pKa = 11.84YY8 pKa = 7.53TGNNSTTSYY17 pKa = 10.59SIPFSYY23 pKa = 10.5RR24 pKa = 11.84ATSDD28 pKa = 3.24LTVTLSGVVTTAYY41 pKa = 9.21TLNSAGTTLTFNTAPAQDD59 pKa = 3.47VAIEE63 pKa = 3.8IRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84TSQGTKK73 pKa = 10.07LVDD76 pKa = 3.56YY77 pKa = 10.83ASGSVLTEE85 pKa = 3.68NDD87 pKa = 3.94LDD89 pKa = 4.06TDD91 pKa = 3.63SDD93 pKa = 3.8QAFFMSQEE101 pKa = 4.56AIDD104 pKa = 4.32DD105 pKa = 4.22AGDD108 pKa = 3.66VIKK111 pKa = 11.09VSNTNFQWDD120 pKa = 3.95VQNKK124 pKa = 9.66RR125 pKa = 11.84LTNVANPVNDD135 pKa = 3.36TDD137 pKa = 4.1AVNKK141 pKa = 9.93QFISTNLPNITTVAGISADD160 pKa = 3.48VTTVAGISSNVTTVANDD177 pKa = 3.35TTNIGTVATNMPSITTVATNINDD200 pKa = 3.93VIKK203 pKa = 11.06VADD206 pKa = 4.73DD207 pKa = 3.71LNEE210 pKa = 4.8AISEE214 pKa = 4.4VEE216 pKa = 4.24TVANDD221 pKa = 3.58LNEE224 pKa = 4.22ATSEE228 pKa = 4.16IEE230 pKa = 4.1VVANNIVNVNTVGTINADD248 pKa = 3.27VTTVANNEE256 pKa = 3.83TDD258 pKa = 3.26IQTLADD264 pKa = 4.24LEE266 pKa = 5.04DD267 pKa = 4.01GTVTTNGLSTLAGLNTEE284 pKa = 3.89IQGVYY289 pKa = 10.52NIRR292 pKa = 11.84TNVTNVDD299 pKa = 3.69TNSANVNLVAGQISPTNNVSAVSAVATEE327 pKa = 3.92IGTLGALGTEE337 pKa = 3.91ITNLNNIRR345 pKa = 11.84TDD347 pKa = 2.9ITGVNNIAPAVTAVNNNSANINTVAGLDD375 pKa = 3.74TEE377 pKa = 4.45ITALGASGTVASINTVASNLASVNNFGEE405 pKa = 4.61VYY407 pKa = 10.0RR408 pKa = 11.84ISATAPTTSLDD419 pKa = 3.8NGDD422 pKa = 3.17MWFDD426 pKa = 3.44TTAGKK431 pKa = 10.42LKK433 pKa = 9.75IWNGSSFDD441 pKa = 4.06LAGSSINGTSARR453 pKa = 11.84FKK455 pKa = 9.34YY456 pKa = 8.74TATASQTTFTGADD469 pKa = 3.49DD470 pKa = 4.13NGNTLAYY477 pKa = 10.26DD478 pKa = 3.44SGFTDD483 pKa = 3.98VYY485 pKa = 11.02LNGVKK490 pKa = 10.07LVNGSDD496 pKa = 3.53YY497 pKa = 10.98TATDD501 pKa = 3.61GSNVVLTTGASVNDD515 pKa = 3.34ILEE518 pKa = 4.12IVGFGTFSVADD529 pKa = 4.37FDD531 pKa = 5.44ASGLTGTINIARR543 pKa = 11.84LADD546 pKa = 3.58NSVTNAKK553 pKa = 9.64LANQSITINGSAVNLGGSVTVGEE576 pKa = 4.52TKK578 pKa = 9.18PTISSISPSTITNAQTSITITGSNFTSVPQVEE610 pKa = 4.76FLNPSTGIWYY620 pKa = 9.82VADD623 pKa = 3.22TVTYY627 pKa = 10.71NNSTSLTVNATLGVDD642 pKa = 3.59GTYY645 pKa = 10.3KK646 pKa = 10.15IRR648 pKa = 11.84IEE650 pKa = 4.26NPDD653 pKa = 3.26GNAVISSTNILTVSDD668 pKa = 3.76APTWTTASGTLGTFAGDD685 pKa = 3.63FSGTLATVVATSDD698 pKa = 3.47SAVTFSEE705 pKa = 4.29VGSNLATANVTLSSGGVLSTTDD727 pKa = 3.79FGGSSTTATTYY738 pKa = 11.13NFTIRR743 pKa = 11.84ATDD746 pKa = 3.77AEE748 pKa = 4.44NQTADD753 pKa = 3.8RR754 pKa = 11.84NFSLTSSFGATGGGQFNN771 pKa = 4.04
Molecular weight: 79.39 kDa
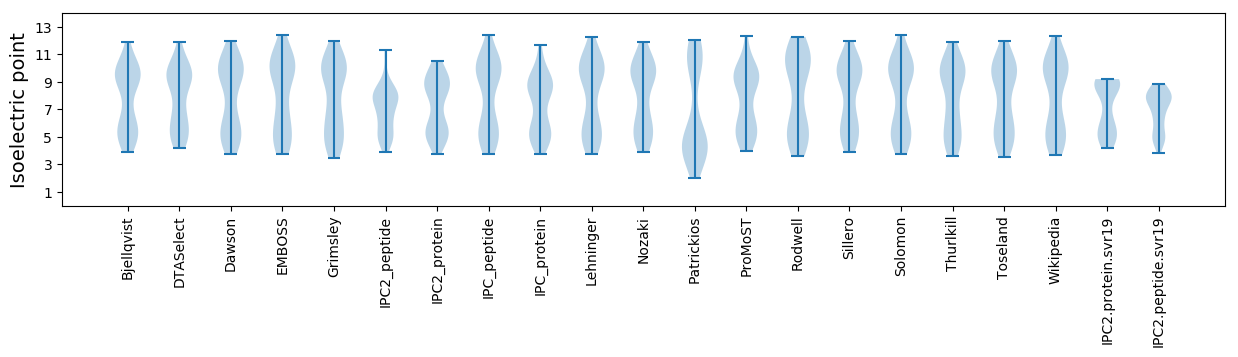
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y1NTQ4|A0A4Y1NTQ4_9CAUD Lysozyme OS=Pelagibacter phage HTVC201P OX=2283024 GN=P201_gp52 PE=4 SV=1
MM1 pKa = 7.74IIYY4 pKa = 9.65GYY6 pKa = 8.41SVKK9 pKa = 8.81TWRR12 pKa = 11.84DD13 pKa = 2.62KK14 pKa = 11.49AIIYY18 pKa = 7.47WRR20 pKa = 11.84NTNKK24 pKa = 10.43KK25 pKa = 10.06LFTLFVLWSIILWAMM40 pKa = 4.03
MM1 pKa = 7.74IIYY4 pKa = 9.65GYY6 pKa = 8.41SVKK9 pKa = 8.81TWRR12 pKa = 11.84DD13 pKa = 2.62KK14 pKa = 11.49AIIYY18 pKa = 7.47WRR20 pKa = 11.84NTNKK24 pKa = 10.43KK25 pKa = 10.06LFTLFVLWSIILWAMM40 pKa = 4.03
Molecular weight: 4.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12862 |
35 |
1247 |
247.3 |
27.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.394 ± 0.381 | 0.879 ± 0.174 |
5.777 ± 0.199 | 6.329 ± 0.482 |
3.965 ± 0.16 | 6.624 ± 0.324 |
1.672 ± 0.203 | 6.655 ± 0.178 |
8.451 ± 0.716 | 7.853 ± 0.392 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.573 ± 0.223 | 6.36 ± 0.413 |
2.939 ± 0.162 | 3.646 ± 0.257 |
3.841 ± 0.305 | 6.912 ± 0.447 |
7.713 ± 0.756 | 5.637 ± 0.364 |
1.291 ± 0.131 | 3.491 ± 0.208 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
