
Streptomyces phage Mischief19
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Woodruffvirus; unclassified Woodruffvirus
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
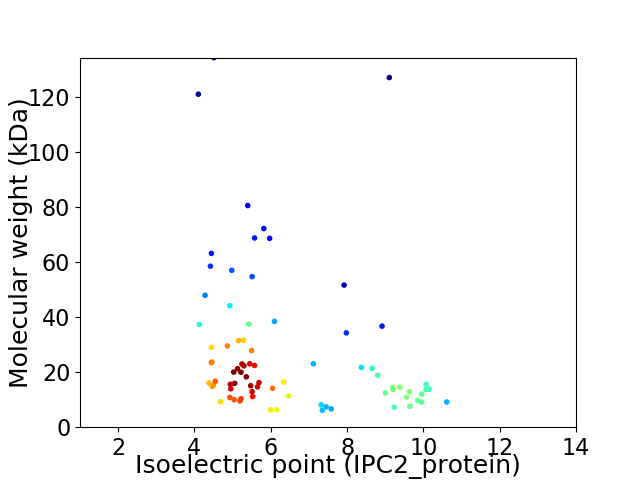
Virtual 2D-PAGE plot for 79 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D6E4B6|A0A4D6E4B6_9CAUD Uncharacterized protein OS=Streptomyces phage Mischief19 OX=2562344 GN=22 PE=4 SV=1
MM1 pKa = 6.63TTPAFNPLPCDD12 pKa = 3.47AGAGGAADD20 pKa = 4.29DD21 pKa = 4.36VEE23 pKa = 4.72SHH25 pKa = 6.7ILCAVDD31 pKa = 3.78ADD33 pKa = 4.12GVVQASALAVYY44 pKa = 9.07TYY46 pKa = 10.52DD47 pKa = 3.51PQGNPVGAPEE57 pKa = 4.17FVDD60 pKa = 4.06PVTGAPFVLPPGTTLQPCPDD80 pKa = 4.5GGCGGPIQFCQTGTTTGPVQHH101 pKa = 7.37PGRR104 pKa = 11.84TYY106 pKa = 10.96DD107 pKa = 2.79ITLPINPGFAVQSLQIDD124 pKa = 4.09SVSQSAGITWEE135 pKa = 3.73VDD137 pKa = 3.29DD138 pKa = 5.08PFGNTFAADD147 pKa = 3.93LKK149 pKa = 11.24AFIEE153 pKa = 4.3GRR155 pKa = 11.84IAGSTVTVTNPNDD168 pKa = 3.68GQIICGTAAPMQVHH182 pKa = 6.19IEE184 pKa = 4.1CLRR187 pKa = 11.84LDD189 pKa = 3.85QNPPNLIEE197 pKa = 4.07LVYY200 pKa = 10.97NAGQDD205 pKa = 4.35LIINPAYY212 pKa = 10.68NEE214 pKa = 4.18NPPLNPPVSQGNYY227 pKa = 9.34GFHH230 pKa = 7.01LLARR234 pKa = 11.84QDD236 pKa = 3.79DD237 pKa = 4.34PGPFPGNQPSGRR249 pKa = 11.84AVCTDD254 pKa = 2.95VANRR258 pKa = 11.84GWEE261 pKa = 4.1TNDD264 pKa = 3.02RR265 pKa = 11.84GRR267 pKa = 11.84TFEE270 pKa = 3.51IWGNNIVTGEE280 pKa = 4.26NVTPTPRR287 pKa = 11.84GTPVQEE293 pKa = 4.16ITSDD297 pKa = 3.82GPPVAGGRR305 pKa = 11.84PSTIWQTFVAPASGNFIIRR324 pKa = 11.84LVHH327 pKa = 5.81GARR330 pKa = 11.84DD331 pKa = 3.31AGEE334 pKa = 3.98VHH336 pKa = 7.73RR337 pKa = 11.84ITLDD341 pKa = 3.37NGDD344 pKa = 4.42TDD346 pKa = 4.15DD347 pKa = 4.04QQNGTLIDD355 pKa = 3.93DD356 pKa = 3.94TTTPGVVTSSTGGPGPWTQFSQTIPLTAGQTYY388 pKa = 8.41TLALSTNNPVGGARR402 pKa = 11.84GGLFTDD408 pKa = 3.71MRR410 pKa = 11.84AFLDD414 pKa = 3.87RR415 pKa = 11.84PDD417 pKa = 3.87RR418 pKa = 11.84RR419 pKa = 11.84ATAVNDD425 pKa = 3.83DD426 pKa = 4.08DD427 pKa = 4.44TCVVTTTEE435 pKa = 4.1TQTTRR440 pKa = 11.84TCSFWQPQCSGGRR453 pKa = 11.84VAGWVKK459 pKa = 10.68VDD461 pKa = 3.36TGLVLSNAEE470 pKa = 4.27FWAQTPTPQCCDD482 pKa = 3.09STGSGDD488 pKa = 4.0GEE490 pKa = 4.21QSSTLSNLAVSDD502 pKa = 3.95IVCATLGGVQQNAVRR517 pKa = 11.84VVVLDD522 pKa = 3.65PSGGQLSEE530 pKa = 4.42TFLGTDD536 pKa = 4.01GARR539 pKa = 11.84IQPASWSPGACTTEE553 pKa = 3.82RR554 pKa = 11.84TLADD558 pKa = 3.65AVLCDD563 pKa = 4.95FGNLALDD570 pKa = 4.0GTPTSFLRR578 pKa = 11.84KK579 pKa = 9.22YY580 pKa = 8.67VQTWSPTVGTSITEE594 pKa = 3.82VRR596 pKa = 11.84DD597 pKa = 3.81FNLSGNGARR606 pKa = 11.84YY607 pKa = 7.55TPLGLVGDD615 pKa = 4.72CSSLRR620 pKa = 11.84PTDD623 pKa = 3.45TEE625 pKa = 4.2TQLVCDD631 pKa = 4.26VQAPIPGQPPASAQIISRR649 pKa = 11.84ITYY652 pKa = 9.81DD653 pKa = 3.08STGAILSQSFTTLDD667 pKa = 3.7GQPYY671 pKa = 8.41TPGGTILQCPNVDD684 pKa = 3.53TEE686 pKa = 4.93LSVHH690 pKa = 6.57CDD692 pKa = 2.8AGNANHH698 pKa = 6.94RR699 pKa = 11.84FVRR702 pKa = 11.84VYY704 pKa = 10.97NRR706 pKa = 11.84VDD708 pKa = 3.31GVQVNFSTVEE718 pKa = 3.78LDD720 pKa = 3.31GVTPYY725 pKa = 10.24TAVGPLVDD733 pKa = 4.33CAAASVSTDD742 pKa = 4.24PIATTGLCLGDD753 pKa = 3.63GTPIAVVTRR762 pKa = 11.84RR763 pKa = 11.84DD764 pKa = 3.37HH765 pKa = 6.45AAGTIITDD773 pKa = 3.33GWVNLLTGTFSAGAPPVGTTACGSSRR799 pKa = 11.84SIEE802 pKa = 3.96LSGVLCDD809 pKa = 3.68VSGGTTVNGLVLVEE823 pKa = 4.02YY824 pKa = 9.86HH825 pKa = 5.93YY826 pKa = 11.41AADD829 pKa = 4.07GSIEE833 pKa = 4.07STRR836 pKa = 11.84LVNATTGATYY846 pKa = 10.85VPVGTITVCPEE857 pKa = 3.6GTSQPDD863 pKa = 2.92RR864 pKa = 11.84DD865 pKa = 4.01MEE867 pKa = 4.43LLCDD871 pKa = 3.74VAANGTVTPFIRR883 pKa = 11.84DD884 pKa = 3.49YY885 pKa = 11.14QRR887 pKa = 11.84NSTGAITGSTSYY899 pKa = 10.12TLGGAAYY906 pKa = 9.74APTGTVGSCSAKK918 pKa = 10.6DD919 pKa = 3.35SEE921 pKa = 4.69TFILCDD927 pKa = 3.32SAAVPVRR934 pKa = 11.84FLRR937 pKa = 11.84TYY939 pKa = 8.74TYY941 pKa = 10.89KK942 pKa = 10.93ADD944 pKa = 3.47GSVAAFTDD952 pKa = 3.78STFAGAAFTPTGAVGTCATTTTPEE976 pKa = 3.67TDD978 pKa = 3.24VQQEE982 pKa = 4.2ILCDD986 pKa = 3.68ANGTQFIRR994 pKa = 11.84RR995 pKa = 11.84WLFNSATGAVTSTVNLTLAGAAFTPVGAVGLCSNCCPVVVGEE1037 pKa = 4.53GCTNTGSGRR1046 pKa = 11.84YY1047 pKa = 8.22VATRR1051 pKa = 11.84AANGTITLQDD1061 pKa = 3.76SVTGATVTQAQIIACPDD1078 pKa = 3.23QPVTVVAEE1086 pKa = 4.11GRR1088 pKa = 11.84IVADD1092 pKa = 5.03ADD1094 pKa = 4.5TPWSPGNDD1102 pKa = 3.07VIGRR1106 pKa = 11.84LTSVTLTVLSGTANVVDD1123 pKa = 4.46GNGTALTGLPAGYY1136 pKa = 7.14TASWDD1141 pKa = 3.75TQDD1144 pKa = 4.87DD1145 pKa = 4.64HH1146 pKa = 6.72NTLTGPTSIDD1156 pKa = 3.16AVGGSTVVVWTRR1168 pKa = 11.84VV1169 pKa = 3.03
MM1 pKa = 6.63TTPAFNPLPCDD12 pKa = 3.47AGAGGAADD20 pKa = 4.29DD21 pKa = 4.36VEE23 pKa = 4.72SHH25 pKa = 6.7ILCAVDD31 pKa = 3.78ADD33 pKa = 4.12GVVQASALAVYY44 pKa = 9.07TYY46 pKa = 10.52DD47 pKa = 3.51PQGNPVGAPEE57 pKa = 4.17FVDD60 pKa = 4.06PVTGAPFVLPPGTTLQPCPDD80 pKa = 4.5GGCGGPIQFCQTGTTTGPVQHH101 pKa = 7.37PGRR104 pKa = 11.84TYY106 pKa = 10.96DD107 pKa = 2.79ITLPINPGFAVQSLQIDD124 pKa = 4.09SVSQSAGITWEE135 pKa = 3.73VDD137 pKa = 3.29DD138 pKa = 5.08PFGNTFAADD147 pKa = 3.93LKK149 pKa = 11.24AFIEE153 pKa = 4.3GRR155 pKa = 11.84IAGSTVTVTNPNDD168 pKa = 3.68GQIICGTAAPMQVHH182 pKa = 6.19IEE184 pKa = 4.1CLRR187 pKa = 11.84LDD189 pKa = 3.85QNPPNLIEE197 pKa = 4.07LVYY200 pKa = 10.97NAGQDD205 pKa = 4.35LIINPAYY212 pKa = 10.68NEE214 pKa = 4.18NPPLNPPVSQGNYY227 pKa = 9.34GFHH230 pKa = 7.01LLARR234 pKa = 11.84QDD236 pKa = 3.79DD237 pKa = 4.34PGPFPGNQPSGRR249 pKa = 11.84AVCTDD254 pKa = 2.95VANRR258 pKa = 11.84GWEE261 pKa = 4.1TNDD264 pKa = 3.02RR265 pKa = 11.84GRR267 pKa = 11.84TFEE270 pKa = 3.51IWGNNIVTGEE280 pKa = 4.26NVTPTPRR287 pKa = 11.84GTPVQEE293 pKa = 4.16ITSDD297 pKa = 3.82GPPVAGGRR305 pKa = 11.84PSTIWQTFVAPASGNFIIRR324 pKa = 11.84LVHH327 pKa = 5.81GARR330 pKa = 11.84DD331 pKa = 3.31AGEE334 pKa = 3.98VHH336 pKa = 7.73RR337 pKa = 11.84ITLDD341 pKa = 3.37NGDD344 pKa = 4.42TDD346 pKa = 4.15DD347 pKa = 4.04QQNGTLIDD355 pKa = 3.93DD356 pKa = 3.94TTTPGVVTSSTGGPGPWTQFSQTIPLTAGQTYY388 pKa = 8.41TLALSTNNPVGGARR402 pKa = 11.84GGLFTDD408 pKa = 3.71MRR410 pKa = 11.84AFLDD414 pKa = 3.87RR415 pKa = 11.84PDD417 pKa = 3.87RR418 pKa = 11.84RR419 pKa = 11.84ATAVNDD425 pKa = 3.83DD426 pKa = 4.08DD427 pKa = 4.44TCVVTTTEE435 pKa = 4.1TQTTRR440 pKa = 11.84TCSFWQPQCSGGRR453 pKa = 11.84VAGWVKK459 pKa = 10.68VDD461 pKa = 3.36TGLVLSNAEE470 pKa = 4.27FWAQTPTPQCCDD482 pKa = 3.09STGSGDD488 pKa = 4.0GEE490 pKa = 4.21QSSTLSNLAVSDD502 pKa = 3.95IVCATLGGVQQNAVRR517 pKa = 11.84VVVLDD522 pKa = 3.65PSGGQLSEE530 pKa = 4.42TFLGTDD536 pKa = 4.01GARR539 pKa = 11.84IQPASWSPGACTTEE553 pKa = 3.82RR554 pKa = 11.84TLADD558 pKa = 3.65AVLCDD563 pKa = 4.95FGNLALDD570 pKa = 4.0GTPTSFLRR578 pKa = 11.84KK579 pKa = 9.22YY580 pKa = 8.67VQTWSPTVGTSITEE594 pKa = 3.82VRR596 pKa = 11.84DD597 pKa = 3.81FNLSGNGARR606 pKa = 11.84YY607 pKa = 7.55TPLGLVGDD615 pKa = 4.72CSSLRR620 pKa = 11.84PTDD623 pKa = 3.45TEE625 pKa = 4.2TQLVCDD631 pKa = 4.26VQAPIPGQPPASAQIISRR649 pKa = 11.84ITYY652 pKa = 9.81DD653 pKa = 3.08STGAILSQSFTTLDD667 pKa = 3.7GQPYY671 pKa = 8.41TPGGTILQCPNVDD684 pKa = 3.53TEE686 pKa = 4.93LSVHH690 pKa = 6.57CDD692 pKa = 2.8AGNANHH698 pKa = 6.94RR699 pKa = 11.84FVRR702 pKa = 11.84VYY704 pKa = 10.97NRR706 pKa = 11.84VDD708 pKa = 3.31GVQVNFSTVEE718 pKa = 3.78LDD720 pKa = 3.31GVTPYY725 pKa = 10.24TAVGPLVDD733 pKa = 4.33CAAASVSTDD742 pKa = 4.24PIATTGLCLGDD753 pKa = 3.63GTPIAVVTRR762 pKa = 11.84RR763 pKa = 11.84DD764 pKa = 3.37HH765 pKa = 6.45AAGTIITDD773 pKa = 3.33GWVNLLTGTFSAGAPPVGTTACGSSRR799 pKa = 11.84SIEE802 pKa = 3.96LSGVLCDD809 pKa = 3.68VSGGTTVNGLVLVEE823 pKa = 4.02YY824 pKa = 9.86HH825 pKa = 5.93YY826 pKa = 11.41AADD829 pKa = 4.07GSIEE833 pKa = 4.07STRR836 pKa = 11.84LVNATTGATYY846 pKa = 10.85VPVGTITVCPEE857 pKa = 3.6GTSQPDD863 pKa = 2.92RR864 pKa = 11.84DD865 pKa = 4.01MEE867 pKa = 4.43LLCDD871 pKa = 3.74VAANGTVTPFIRR883 pKa = 11.84DD884 pKa = 3.49YY885 pKa = 11.14QRR887 pKa = 11.84NSTGAITGSTSYY899 pKa = 10.12TLGGAAYY906 pKa = 9.74APTGTVGSCSAKK918 pKa = 10.6DD919 pKa = 3.35SEE921 pKa = 4.69TFILCDD927 pKa = 3.32SAAVPVRR934 pKa = 11.84FLRR937 pKa = 11.84TYY939 pKa = 8.74TYY941 pKa = 10.89KK942 pKa = 10.93ADD944 pKa = 3.47GSVAAFTDD952 pKa = 3.78STFAGAAFTPTGAVGTCATTTTPEE976 pKa = 3.67TDD978 pKa = 3.24VQQEE982 pKa = 4.2ILCDD986 pKa = 3.68ANGTQFIRR994 pKa = 11.84RR995 pKa = 11.84WLFNSATGAVTSTVNLTLAGAAFTPVGAVGLCSNCCPVVVGEE1037 pKa = 4.53GCTNTGSGRR1046 pKa = 11.84YY1047 pKa = 8.22VATRR1051 pKa = 11.84AANGTITLQDD1061 pKa = 3.76SVTGATVTQAQIIACPDD1078 pKa = 3.23QPVTVVAEE1086 pKa = 4.11GRR1088 pKa = 11.84IVADD1092 pKa = 5.03ADD1094 pKa = 4.5TPWSPGNDD1102 pKa = 3.07VIGRR1106 pKa = 11.84LTSVTLTVLSGTANVVDD1123 pKa = 4.46GNGTALTGLPAGYY1136 pKa = 7.14TASWDD1141 pKa = 3.75TQDD1144 pKa = 4.87DD1145 pKa = 4.64HH1146 pKa = 6.72NTLTGPTSIDD1156 pKa = 3.16AVGGSTVVVWTRR1168 pKa = 11.84VV1169 pKa = 3.03
Molecular weight: 120.86 kDa
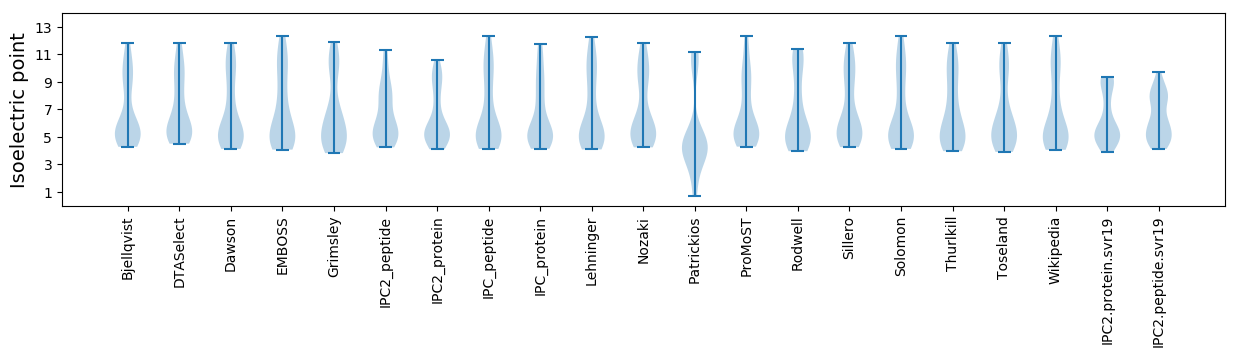
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D6E4L5|A0A4D6E4L5_9CAUD Uncharacterized protein OS=Streptomyces phage Mischief19 OX=2562344 GN=71 PE=4 SV=1
MM1 pKa = 7.58TNTTATKK8 pKa = 8.54VTAAEE13 pKa = 4.21LVAGTIVWGRR23 pKa = 11.84PVRR26 pKa = 11.84EE27 pKa = 4.23GKK29 pKa = 9.96AQSRR33 pKa = 11.84RR34 pKa = 11.84KK35 pKa = 10.23GLVLGSFGDD44 pKa = 3.72SAVIVWWYY52 pKa = 11.14GEE54 pKa = 4.26GAASGSTTTMAFPRR68 pKa = 11.84EE69 pKa = 4.11LTRR72 pKa = 11.84DD73 pKa = 2.8GDD75 pKa = 4.06MFDD78 pKa = 3.55LSARR82 pKa = 11.84VARR85 pKa = 11.84KK86 pKa = 8.51LAKK89 pKa = 9.63GCYY92 pKa = 8.28GFEE95 pKa = 4.15RR96 pKa = 11.84AASVGWSLTRR106 pKa = 11.84HH107 pKa = 5.9AARR110 pKa = 11.84MASLGASQPVRR121 pKa = 3.42
MM1 pKa = 7.58TNTTATKK8 pKa = 8.54VTAAEE13 pKa = 4.21LVAGTIVWGRR23 pKa = 11.84PVRR26 pKa = 11.84EE27 pKa = 4.23GKK29 pKa = 9.96AQSRR33 pKa = 11.84RR34 pKa = 11.84KK35 pKa = 10.23GLVLGSFGDD44 pKa = 3.72SAVIVWWYY52 pKa = 11.14GEE54 pKa = 4.26GAASGSTTTMAFPRR68 pKa = 11.84EE69 pKa = 4.11LTRR72 pKa = 11.84DD73 pKa = 2.8GDD75 pKa = 4.06MFDD78 pKa = 3.55LSARR82 pKa = 11.84VARR85 pKa = 11.84KK86 pKa = 8.51LAKK89 pKa = 9.63GCYY92 pKa = 8.28GFEE95 pKa = 4.15RR96 pKa = 11.84AASVGWSLTRR106 pKa = 11.84HH107 pKa = 5.9AARR110 pKa = 11.84MASLGASQPVRR121 pKa = 3.42
Molecular weight: 12.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
20485 |
57 |
1279 |
259.3 |
27.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.41 ± 0.574 | 1.679 ± 0.203 |
6.092 ± 0.242 | 4.926 ± 0.312 |
2.504 ± 0.183 | 9.666 ± 0.401 |
1.816 ± 0.17 | 3.691 ± 0.242 |
3.056 ± 0.302 | 8.352 ± 0.28 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.884 ± 0.14 | 3.373 ± 0.241 |
5.297 ± 0.271 | 3.061 ± 0.141 |
6.375 ± 0.336 | 5.648 ± 0.25 |
7.508 ± 0.448 | 8.05 ± 0.162 |
1.425 ± 0.13 | 2.187 ± 0.154 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
