
Lachnospiraceae bacterium A4
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; unclassified Lachnospiraceae
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
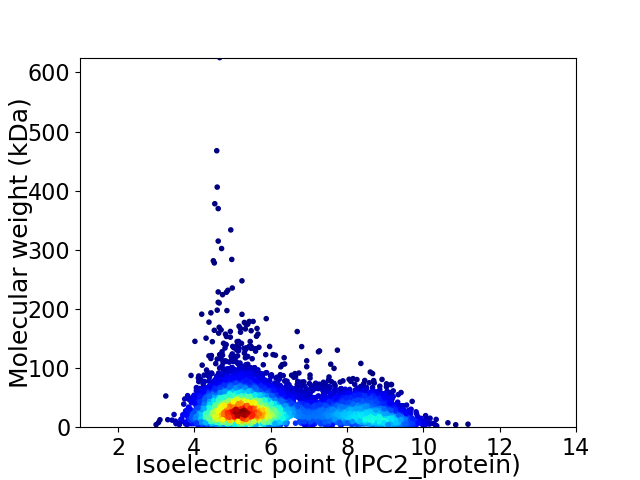
Virtual 2D-PAGE plot for 6440 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9JA65|R9JA65_9FIRM Uncharacterized protein (Fragment) OS=Lachnospiraceae bacterium A4 OX=397291 GN=C804_02945 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 10.38LKK4 pKa = 10.68RR5 pKa = 11.84LMALGMAAVMTMSLTACGGSGDD27 pKa = 4.09TPSTNSNTASNNNDD41 pKa = 2.96SNAAADD47 pKa = 3.85NDD49 pKa = 4.3AAADD53 pKa = 3.94DD54 pKa = 5.04ANAADD59 pKa = 5.74DD60 pKa = 4.81SAADD64 pKa = 4.1DD65 pKa = 4.75ANAADD70 pKa = 4.68DD71 pKa = 3.93SAAEE75 pKa = 4.39GGLTYY80 pKa = 10.98GSITLGEE87 pKa = 5.02DD88 pKa = 3.51YY89 pKa = 11.04TDD91 pKa = 3.44LTASIKK97 pKa = 10.28FIHH100 pKa = 6.62HH101 pKa = 5.46KK102 pKa = 8.62TDD104 pKa = 3.48RR105 pKa = 11.84EE106 pKa = 4.12QDD108 pKa = 3.12GSMAKK113 pKa = 10.45YY114 pKa = 10.04IEE116 pKa = 4.86AFNQEE121 pKa = 4.26YY122 pKa = 10.3PNITVEE128 pKa = 4.3TEE130 pKa = 4.01GITDD134 pKa = 3.73YY135 pKa = 11.58AQTALLRR142 pKa = 11.84LPTGEE147 pKa = 3.76AWGDD151 pKa = 3.33VMFIPSIDD159 pKa = 3.56MVQLADD165 pKa = 3.54YY166 pKa = 7.29FIPYY170 pKa = 8.26GTVDD174 pKa = 3.32EE175 pKa = 4.29MSEE178 pKa = 4.19YY179 pKa = 11.45VNFADD184 pKa = 3.26QWKK187 pKa = 10.52FEE189 pKa = 4.36GNCYY193 pKa = 9.92GVGYY197 pKa = 9.11MGNAQGILYY206 pKa = 9.0NKK208 pKa = 9.97KK209 pKa = 8.58VFADD213 pKa = 3.46AGVTEE218 pKa = 4.73LPKK221 pKa = 10.73TPTEE225 pKa = 4.86FIAALQQIKK234 pKa = 10.73DD235 pKa = 3.68NTDD238 pKa = 4.57AIPLYY243 pKa = 10.26TNYY246 pKa = 10.71AAGWTMGGQWDD257 pKa = 4.59AYY259 pKa = 10.42LGAITTGDD267 pKa = 3.96TTWLNQKK274 pKa = 9.03FAHH277 pKa = 5.95SAEE280 pKa = 4.13PFKK283 pKa = 11.55DD284 pKa = 3.43NGDD287 pKa = 3.5EE288 pKa = 4.01TGAYY292 pKa = 9.86ALYY295 pKa = 10.22KK296 pKa = 10.11ILYY299 pKa = 8.87DD300 pKa = 3.67AVANGLTEE308 pKa = 5.37DD309 pKa = 5.44DD310 pKa = 4.16YY311 pKa = 10.86TTTDD315 pKa = 2.86WEE317 pKa = 4.93GCKK320 pKa = 10.76GMINRR325 pKa = 11.84GEE327 pKa = 4.23IGTMVLGSWAVAQMKK342 pKa = 9.84EE343 pKa = 3.68ADD345 pKa = 4.06ANGGDD350 pKa = 3.7IGYY353 pKa = 8.11MPFPISVGGKK363 pKa = 8.89QYY365 pKa = 10.33ATAGPDD371 pKa = 3.12YY372 pKa = 10.71CYY374 pKa = 10.73GINKK378 pKa = 9.28EE379 pKa = 4.15SSADD383 pKa = 3.55NQAAAMVFVKK393 pKa = 10.03WMVEE397 pKa = 3.68KK398 pKa = 10.94SGWCDD403 pKa = 3.25NEE405 pKa = 3.8GGYY408 pKa = 9.62PISKK412 pKa = 8.26TAPTSFVFDD421 pKa = 3.36GVEE424 pKa = 4.12VVNNEE429 pKa = 3.86AAPDD433 pKa = 3.93DD434 pKa = 4.5EE435 pKa = 6.91ADD437 pKa = 5.75LMNQMNAEE445 pKa = 4.16CEE447 pKa = 4.3LNFNNGGDD455 pKa = 3.31RR456 pKa = 11.84KK457 pKa = 9.72VQEE460 pKa = 4.22IVEE463 pKa = 4.12AAATGSMTYY472 pKa = 10.79DD473 pKa = 3.24EE474 pKa = 6.06IMANWTALWNDD485 pKa = 3.5AQSSLGVDD493 pKa = 3.42VTEE496 pKa = 4.38
MM1 pKa = 7.44KK2 pKa = 10.38LKK4 pKa = 10.68RR5 pKa = 11.84LMALGMAAVMTMSLTACGGSGDD27 pKa = 4.09TPSTNSNTASNNNDD41 pKa = 2.96SNAAADD47 pKa = 3.85NDD49 pKa = 4.3AAADD53 pKa = 3.94DD54 pKa = 5.04ANAADD59 pKa = 5.74DD60 pKa = 4.81SAADD64 pKa = 4.1DD65 pKa = 4.75ANAADD70 pKa = 4.68DD71 pKa = 3.93SAAEE75 pKa = 4.39GGLTYY80 pKa = 10.98GSITLGEE87 pKa = 5.02DD88 pKa = 3.51YY89 pKa = 11.04TDD91 pKa = 3.44LTASIKK97 pKa = 10.28FIHH100 pKa = 6.62HH101 pKa = 5.46KK102 pKa = 8.62TDD104 pKa = 3.48RR105 pKa = 11.84EE106 pKa = 4.12QDD108 pKa = 3.12GSMAKK113 pKa = 10.45YY114 pKa = 10.04IEE116 pKa = 4.86AFNQEE121 pKa = 4.26YY122 pKa = 10.3PNITVEE128 pKa = 4.3TEE130 pKa = 4.01GITDD134 pKa = 3.73YY135 pKa = 11.58AQTALLRR142 pKa = 11.84LPTGEE147 pKa = 3.76AWGDD151 pKa = 3.33VMFIPSIDD159 pKa = 3.56MVQLADD165 pKa = 3.54YY166 pKa = 7.29FIPYY170 pKa = 8.26GTVDD174 pKa = 3.32EE175 pKa = 4.29MSEE178 pKa = 4.19YY179 pKa = 11.45VNFADD184 pKa = 3.26QWKK187 pKa = 10.52FEE189 pKa = 4.36GNCYY193 pKa = 9.92GVGYY197 pKa = 9.11MGNAQGILYY206 pKa = 9.0NKK208 pKa = 9.97KK209 pKa = 8.58VFADD213 pKa = 3.46AGVTEE218 pKa = 4.73LPKK221 pKa = 10.73TPTEE225 pKa = 4.86FIAALQQIKK234 pKa = 10.73DD235 pKa = 3.68NTDD238 pKa = 4.57AIPLYY243 pKa = 10.26TNYY246 pKa = 10.71AAGWTMGGQWDD257 pKa = 4.59AYY259 pKa = 10.42LGAITTGDD267 pKa = 3.96TTWLNQKK274 pKa = 9.03FAHH277 pKa = 5.95SAEE280 pKa = 4.13PFKK283 pKa = 11.55DD284 pKa = 3.43NGDD287 pKa = 3.5EE288 pKa = 4.01TGAYY292 pKa = 9.86ALYY295 pKa = 10.22KK296 pKa = 10.11ILYY299 pKa = 8.87DD300 pKa = 3.67AVANGLTEE308 pKa = 5.37DD309 pKa = 5.44DD310 pKa = 4.16YY311 pKa = 10.86TTTDD315 pKa = 2.86WEE317 pKa = 4.93GCKK320 pKa = 10.76GMINRR325 pKa = 11.84GEE327 pKa = 4.23IGTMVLGSWAVAQMKK342 pKa = 9.84EE343 pKa = 3.68ADD345 pKa = 4.06ANGGDD350 pKa = 3.7IGYY353 pKa = 8.11MPFPISVGGKK363 pKa = 8.89QYY365 pKa = 10.33ATAGPDD371 pKa = 3.12YY372 pKa = 10.71CYY374 pKa = 10.73GINKK378 pKa = 9.28EE379 pKa = 4.15SSADD383 pKa = 3.55NQAAAMVFVKK393 pKa = 10.03WMVEE397 pKa = 3.68KK398 pKa = 10.94SGWCDD403 pKa = 3.25NEE405 pKa = 3.8GGYY408 pKa = 9.62PISKK412 pKa = 8.26TAPTSFVFDD421 pKa = 3.36GVEE424 pKa = 4.12VVNNEE429 pKa = 3.86AAPDD433 pKa = 3.93DD434 pKa = 4.5EE435 pKa = 6.91ADD437 pKa = 5.75LMNQMNAEE445 pKa = 4.16CEE447 pKa = 4.3LNFNNGGDD455 pKa = 3.31RR456 pKa = 11.84KK457 pKa = 9.72VQEE460 pKa = 4.22IVEE463 pKa = 4.12AAATGSMTYY472 pKa = 10.79DD473 pKa = 3.24EE474 pKa = 6.06IMANWTALWNDD485 pKa = 3.5AQSSLGVDD493 pKa = 3.42VTEE496 pKa = 4.38
Molecular weight: 53.35 kDa
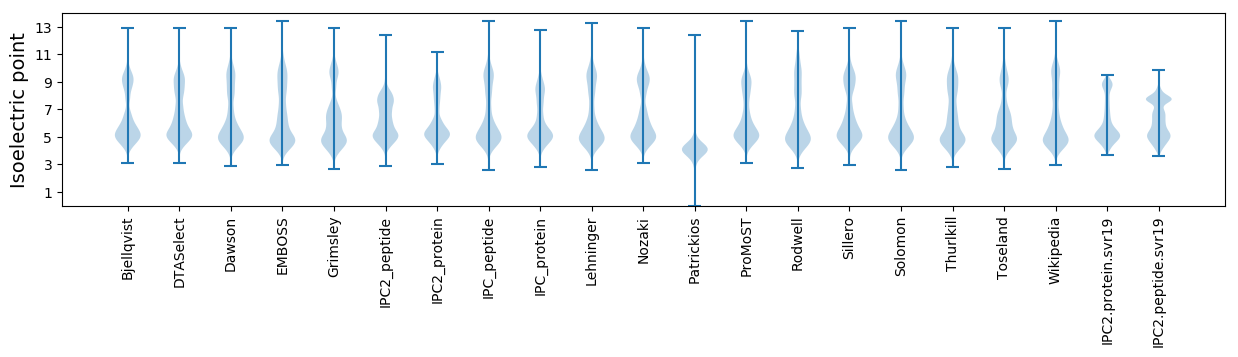
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R9JUY2|R9JUY2_9FIRM Uncharacterized protein OS=Lachnospiraceae bacterium A4 OX=397291 GN=C804_00046 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1943585 |
20 |
5621 |
301.8 |
34.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.351 ± 0.034 | 1.591 ± 0.014 |
5.774 ± 0.027 | 7.793 ± 0.037 |
4.099 ± 0.023 | 6.464 ± 0.029 |
1.77 ± 0.014 | 7.107 ± 0.032 |
6.796 ± 0.029 | 8.818 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.046 ± 0.019 | 4.613 ± 0.024 |
3.025 ± 0.017 | 3.673 ± 0.022 |
4.754 ± 0.026 | 5.835 ± 0.026 |
5.379 ± 0.035 | 6.462 ± 0.027 |
1.021 ± 0.011 | 4.627 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
