
Rhodobacter sp. SW2
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Rhodobacter; unclassified Rhodobacter
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
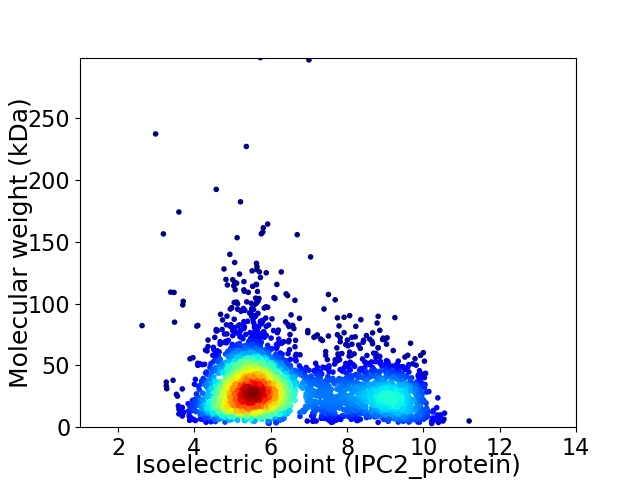
Virtual 2D-PAGE plot for 3405 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
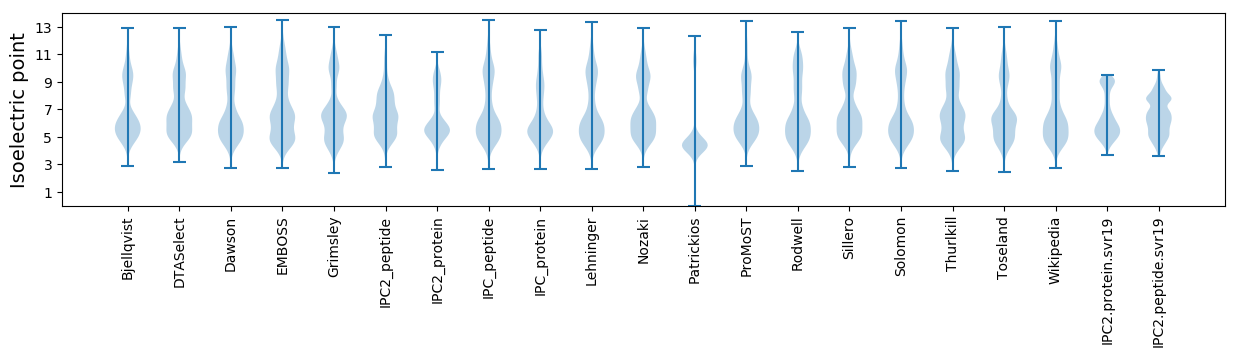
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C8S4P7|C8S4P7_9RHOB Uncharacterized protein OS=Rhodobacter sp. SW2 OX=371731 GN=Rsw2DRAFT_3025 PE=4 SV=1
MM1 pKa = 6.9TQGLLALIMLASVLIGGTGQAQAQDD26 pKa = 4.2VNWVLSLSDD35 pKa = 3.82AGSDD39 pKa = 3.35PVAAGGTIAYY49 pKa = 7.37TMSVTNDD56 pKa = 3.29GFDD59 pKa = 3.57AAPPTSITLQIPANTRR75 pKa = 11.84FTGATGSITGCAPTPSIGPSSVTCNVPPLAPSAVATLSANLLTSVSGDD123 pKa = 3.46VFFSASVPPVGDD135 pKa = 3.61TDD137 pKa = 4.15PADD140 pKa = 3.69NDD142 pKa = 3.59LTEE145 pKa = 4.09QTTITAGADD154 pKa = 3.02VGLVLRR160 pKa = 11.84GPATASAGEE169 pKa = 4.19VISYY173 pKa = 10.62SFAATNNGPDD183 pKa = 3.62PVTDD187 pKa = 4.5LVLQFPIPSGVTNVVAPAGCVLTATTYY214 pKa = 11.25DD215 pKa = 3.63CTIPGPVAVGASVNVDD231 pKa = 3.56FTGQISAASGSTVTAIGSVGAGNPSDD257 pKa = 5.58PIDD260 pKa = 4.51SNDD263 pKa = 3.47NASLNTSVTAGSDD276 pKa = 3.17LTIAKK281 pKa = 9.65SRR283 pKa = 11.84SPSGSLLVGNPAVFTLTPSYY303 pKa = 10.67TGDD306 pKa = 3.56SPNGIVVTDD315 pKa = 4.97SIPANYY321 pKa = 9.47TIDD324 pKa = 3.61TVTAPGWICTVTGQVVEE341 pKa = 4.53CSRR344 pKa = 11.84TTGSGAGANVPLGPISIATTVATPGTPTNTTSITAAGPDD383 pKa = 3.84DD384 pKa = 4.43PVPGNNSATDD394 pKa = 3.55GGATIEE400 pKa = 4.33EE401 pKa = 4.56PVVDD405 pKa = 4.68LQANKK410 pKa = 10.33SGPFPALVVVGNSYY424 pKa = 11.56DD425 pKa = 3.81FTISTSNVGNAAFFGTLEE443 pKa = 4.18MTDD446 pKa = 3.69TLPAGLTVTAYY457 pKa = 10.49AEE459 pKa = 4.28NGWTCTPAVSVVGPASILCSRR480 pKa = 11.84VYY482 pKa = 9.82TAGAPLAAGATTPSVTLTTTATTAGSVVNTMSVLSPDD519 pKa = 3.48SNLPEE524 pKa = 5.67DD525 pKa = 4.14PLKK528 pKa = 11.24DD529 pKa = 4.33PLILDD534 pKa = 4.12NNTTTYY540 pKa = 10.96AVSSSEE546 pKa = 4.41GPDD549 pKa = 3.25SADD552 pKa = 2.74ISVIKK557 pKa = 10.59SATLASLNAGEE568 pKa = 4.47VQTFTIEE575 pKa = 4.15VVNNGPVVSTDD586 pKa = 3.52VEE588 pKa = 4.43MTDD591 pKa = 3.84NLQGLINSSVGAAGAGYY608 pKa = 10.11ISDD611 pKa = 3.99SFTTGAAGGISCSTAATGGTSRR633 pKa = 11.84EE634 pKa = 4.08LSCVIDD640 pKa = 3.64TLPICTAGVDD650 pKa = 3.87CPVFTVQVRR659 pKa = 11.84PGGDD663 pKa = 2.68GGARR667 pKa = 11.84TNTASAISQTVPDD680 pKa = 4.47SNLGNNADD688 pKa = 3.88SAPFTLAPRR697 pKa = 11.84VDD699 pKa = 3.85VAVSKK704 pKa = 11.06ADD706 pKa = 3.67SPDD709 pKa = 3.43PAAAGQNLTYY719 pKa = 10.85VITAQNLANGLSTATGVTITDD740 pKa = 3.82TLPLNVTFVSASPSSGTCGTTPPANSTTTAGSRR773 pKa = 11.84TVSCVLGSIGNGAQQTVTVIVRR795 pKa = 11.84PNLITRR801 pKa = 11.84NTTLQNDD808 pKa = 3.78VAYY811 pKa = 10.44SGAVPDD817 pKa = 4.59IEE819 pKa = 4.72TGNDD823 pKa = 3.05TASATTFVSPPRR835 pKa = 11.84IDD837 pKa = 5.64LIVNKK842 pKa = 10.05DD843 pKa = 3.61DD844 pKa = 4.05TVDD847 pKa = 3.82PLPIGDD853 pKa = 3.87DD854 pKa = 3.22TTYY857 pKa = 11.35LVTVANLGPSSAEE870 pKa = 3.71NVLVTDD876 pKa = 3.61TMPPSRR882 pKa = 11.84LTYY885 pKa = 9.48QSHH888 pKa = 5.78VATGATCNSVPALDD902 pKa = 4.94SLGQTLACTFPVIPAGEE919 pKa = 4.26SRR921 pKa = 11.84TISITARR928 pKa = 11.84GVAKK932 pKa = 10.43GVGLNSVSVSSAEE945 pKa = 3.91TALGYY950 pKa = 10.33EE951 pKa = 4.55PNTGNNSTTEE961 pKa = 3.85QTSIRR966 pKa = 11.84TRR968 pKa = 11.84ADD970 pKa = 2.98MEE972 pKa = 4.6VASKK976 pKa = 8.95TATPATVNLRR986 pKa = 11.84DD987 pKa = 3.74PFSYY991 pKa = 10.85LIVVEE996 pKa = 4.45NNTGTGLAEE1005 pKa = 4.52ADD1007 pKa = 3.72DD1008 pKa = 4.3VVVSDD1013 pKa = 4.34TLPSGMQLSGAPTVSVISGTTTTSTCTGASGSTSFTCALGTVSSGAVVHH1062 pKa = 5.18VTVPARR1068 pKa = 11.84IVTVTALGQTFTNTASVSTSSLDD1091 pKa = 3.48VLPGNNSNSGPVVANSSSLSGTVFRR1116 pKa = 11.84DD1117 pKa = 4.52FAANGTLDD1125 pKa = 3.67GSDD1128 pKa = 3.18TGIVGVTMTLTGTALDD1144 pKa = 3.89GTPVSRR1150 pKa = 11.84TVTTGAGGAYY1160 pKa = 10.17AFDD1163 pKa = 4.47FLPEE1167 pKa = 3.98GTYY1170 pKa = 10.47SISQGAISEE1179 pKa = 4.62SYY1181 pKa = 8.68LTNGATAAGPGGGTPAPTQITAIPLLPATASPGYY1215 pKa = 9.67IFPKK1219 pKa = 9.86VPQARR1224 pKa = 11.84VAIAKK1229 pKa = 9.27AVQAGPMINADD1240 pKa = 2.78GSYY1243 pKa = 10.84NVTFRR1248 pKa = 11.84LRR1250 pKa = 11.84VQNPSLEE1257 pKa = 4.03GLTNVAVTDD1266 pKa = 4.32PLEE1269 pKa = 4.37GAAPLFGTYY1278 pKa = 9.18VALAAPTVPGTYY1290 pKa = 10.0TMVAAPSGSCGGLNGGFTGAGPATVASGFAIAAGATCTIDD1330 pKa = 3.16LQLRR1334 pKa = 11.84VQPTQPLPPLTGAGRR1349 pKa = 11.84YY1350 pKa = 8.87LNQATVDD1357 pKa = 3.67AQGVLSGQTSASNPQLTDD1375 pKa = 3.33LSDD1378 pKa = 3.49NGTSADD1384 pKa = 3.44ANGNGRR1390 pKa = 11.84GNEE1393 pKa = 3.97AGEE1396 pKa = 4.14NDD1398 pKa = 4.35PTPVTPVYY1406 pKa = 10.76NPAITLTKK1414 pKa = 10.58AIDD1417 pKa = 3.93LSVLPSPIAADD1428 pKa = 3.62SPISFTFTVTNSGNITLTDD1447 pKa = 3.47VTLSDD1452 pKa = 3.81ALAGAVVTGGPITLAPGQVDD1472 pKa = 3.73STSFTATYY1480 pKa = 9.24PLSAADD1486 pKa = 3.68LTANTLSNTATATGTWGLDD1505 pKa = 3.16AGGNPQKK1512 pKa = 10.96VSDD1515 pKa = 4.77PDD1517 pKa = 3.68TVATSFADD1525 pKa = 3.21IALVKK1530 pKa = 10.55AADD1533 pKa = 3.68ASLLSVPPQVGDD1545 pKa = 3.6IISYY1549 pKa = 10.83DD1550 pKa = 3.53FAVTNTGNVPLTNVTLSDD1568 pKa = 3.43ILTDD1572 pKa = 3.59AVLTGGPIPSLAVGATDD1589 pKa = 3.8SSTFTATYY1597 pKa = 10.97ALTQADD1603 pKa = 3.42IDD1605 pKa = 3.77AGEE1608 pKa = 4.36VINRR1612 pKa = 11.84ATADD1616 pKa = 3.56GVYY1619 pKa = 9.34GTDD1622 pKa = 4.69DD1623 pKa = 3.9GGDD1626 pKa = 3.95PISVQDD1632 pKa = 3.54EE1633 pKa = 4.16SGANVTEE1640 pKa = 4.73DD1641 pKa = 3.48ADD1643 pKa = 4.19TVVPLTQAPRR1653 pKa = 11.84IEE1655 pKa = 4.85LIKK1658 pKa = 10.79SLSSLVDD1665 pKa = 3.27TTGDD1669 pKa = 3.68GQIGAGDD1676 pKa = 3.52TANYY1680 pKa = 9.57GFTVTNTGNVALAGVTVSDD1699 pKa = 4.34LLVSVSGGPVSLAIGATDD1717 pKa = 3.43STSFTAAYY1725 pKa = 10.45VLTQADD1731 pKa = 3.37VDD1733 pKa = 3.69RR1734 pKa = 11.84GYY1736 pKa = 10.97VQNTATATGAAVTSSGDD1753 pKa = 3.79PILDD1757 pKa = 3.35
MM1 pKa = 6.9TQGLLALIMLASVLIGGTGQAQAQDD26 pKa = 4.2VNWVLSLSDD35 pKa = 3.82AGSDD39 pKa = 3.35PVAAGGTIAYY49 pKa = 7.37TMSVTNDD56 pKa = 3.29GFDD59 pKa = 3.57AAPPTSITLQIPANTRR75 pKa = 11.84FTGATGSITGCAPTPSIGPSSVTCNVPPLAPSAVATLSANLLTSVSGDD123 pKa = 3.46VFFSASVPPVGDD135 pKa = 3.61TDD137 pKa = 4.15PADD140 pKa = 3.69NDD142 pKa = 3.59LTEE145 pKa = 4.09QTTITAGADD154 pKa = 3.02VGLVLRR160 pKa = 11.84GPATASAGEE169 pKa = 4.19VISYY173 pKa = 10.62SFAATNNGPDD183 pKa = 3.62PVTDD187 pKa = 4.5LVLQFPIPSGVTNVVAPAGCVLTATTYY214 pKa = 11.25DD215 pKa = 3.63CTIPGPVAVGASVNVDD231 pKa = 3.56FTGQISAASGSTVTAIGSVGAGNPSDD257 pKa = 5.58PIDD260 pKa = 4.51SNDD263 pKa = 3.47NASLNTSVTAGSDD276 pKa = 3.17LTIAKK281 pKa = 9.65SRR283 pKa = 11.84SPSGSLLVGNPAVFTLTPSYY303 pKa = 10.67TGDD306 pKa = 3.56SPNGIVVTDD315 pKa = 4.97SIPANYY321 pKa = 9.47TIDD324 pKa = 3.61TVTAPGWICTVTGQVVEE341 pKa = 4.53CSRR344 pKa = 11.84TTGSGAGANVPLGPISIATTVATPGTPTNTTSITAAGPDD383 pKa = 3.84DD384 pKa = 4.43PVPGNNSATDD394 pKa = 3.55GGATIEE400 pKa = 4.33EE401 pKa = 4.56PVVDD405 pKa = 4.68LQANKK410 pKa = 10.33SGPFPALVVVGNSYY424 pKa = 11.56DD425 pKa = 3.81FTISTSNVGNAAFFGTLEE443 pKa = 4.18MTDD446 pKa = 3.69TLPAGLTVTAYY457 pKa = 10.49AEE459 pKa = 4.28NGWTCTPAVSVVGPASILCSRR480 pKa = 11.84VYY482 pKa = 9.82TAGAPLAAGATTPSVTLTTTATTAGSVVNTMSVLSPDD519 pKa = 3.48SNLPEE524 pKa = 5.67DD525 pKa = 4.14PLKK528 pKa = 11.24DD529 pKa = 4.33PLILDD534 pKa = 4.12NNTTTYY540 pKa = 10.96AVSSSEE546 pKa = 4.41GPDD549 pKa = 3.25SADD552 pKa = 2.74ISVIKK557 pKa = 10.59SATLASLNAGEE568 pKa = 4.47VQTFTIEE575 pKa = 4.15VVNNGPVVSTDD586 pKa = 3.52VEE588 pKa = 4.43MTDD591 pKa = 3.84NLQGLINSSVGAAGAGYY608 pKa = 10.11ISDD611 pKa = 3.99SFTTGAAGGISCSTAATGGTSRR633 pKa = 11.84EE634 pKa = 4.08LSCVIDD640 pKa = 3.64TLPICTAGVDD650 pKa = 3.87CPVFTVQVRR659 pKa = 11.84PGGDD663 pKa = 2.68GGARR667 pKa = 11.84TNTASAISQTVPDD680 pKa = 4.47SNLGNNADD688 pKa = 3.88SAPFTLAPRR697 pKa = 11.84VDD699 pKa = 3.85VAVSKK704 pKa = 11.06ADD706 pKa = 3.67SPDD709 pKa = 3.43PAAAGQNLTYY719 pKa = 10.85VITAQNLANGLSTATGVTITDD740 pKa = 3.82TLPLNVTFVSASPSSGTCGTTPPANSTTTAGSRR773 pKa = 11.84TVSCVLGSIGNGAQQTVTVIVRR795 pKa = 11.84PNLITRR801 pKa = 11.84NTTLQNDD808 pKa = 3.78VAYY811 pKa = 10.44SGAVPDD817 pKa = 4.59IEE819 pKa = 4.72TGNDD823 pKa = 3.05TASATTFVSPPRR835 pKa = 11.84IDD837 pKa = 5.64LIVNKK842 pKa = 10.05DD843 pKa = 3.61DD844 pKa = 4.05TVDD847 pKa = 3.82PLPIGDD853 pKa = 3.87DD854 pKa = 3.22TTYY857 pKa = 11.35LVTVANLGPSSAEE870 pKa = 3.71NVLVTDD876 pKa = 3.61TMPPSRR882 pKa = 11.84LTYY885 pKa = 9.48QSHH888 pKa = 5.78VATGATCNSVPALDD902 pKa = 4.94SLGQTLACTFPVIPAGEE919 pKa = 4.26SRR921 pKa = 11.84TISITARR928 pKa = 11.84GVAKK932 pKa = 10.43GVGLNSVSVSSAEE945 pKa = 3.91TALGYY950 pKa = 10.33EE951 pKa = 4.55PNTGNNSTTEE961 pKa = 3.85QTSIRR966 pKa = 11.84TRR968 pKa = 11.84ADD970 pKa = 2.98MEE972 pKa = 4.6VASKK976 pKa = 8.95TATPATVNLRR986 pKa = 11.84DD987 pKa = 3.74PFSYY991 pKa = 10.85LIVVEE996 pKa = 4.45NNTGTGLAEE1005 pKa = 4.52ADD1007 pKa = 3.72DD1008 pKa = 4.3VVVSDD1013 pKa = 4.34TLPSGMQLSGAPTVSVISGTTTTSTCTGASGSTSFTCALGTVSSGAVVHH1062 pKa = 5.18VTVPARR1068 pKa = 11.84IVTVTALGQTFTNTASVSTSSLDD1091 pKa = 3.48VLPGNNSNSGPVVANSSSLSGTVFRR1116 pKa = 11.84DD1117 pKa = 4.52FAANGTLDD1125 pKa = 3.67GSDD1128 pKa = 3.18TGIVGVTMTLTGTALDD1144 pKa = 3.89GTPVSRR1150 pKa = 11.84TVTTGAGGAYY1160 pKa = 10.17AFDD1163 pKa = 4.47FLPEE1167 pKa = 3.98GTYY1170 pKa = 10.47SISQGAISEE1179 pKa = 4.62SYY1181 pKa = 8.68LTNGATAAGPGGGTPAPTQITAIPLLPATASPGYY1215 pKa = 9.67IFPKK1219 pKa = 9.86VPQARR1224 pKa = 11.84VAIAKK1229 pKa = 9.27AVQAGPMINADD1240 pKa = 2.78GSYY1243 pKa = 10.84NVTFRR1248 pKa = 11.84LRR1250 pKa = 11.84VQNPSLEE1257 pKa = 4.03GLTNVAVTDD1266 pKa = 4.32PLEE1269 pKa = 4.37GAAPLFGTYY1278 pKa = 9.18VALAAPTVPGTYY1290 pKa = 10.0TMVAAPSGSCGGLNGGFTGAGPATVASGFAIAAGATCTIDD1330 pKa = 3.16LQLRR1334 pKa = 11.84VQPTQPLPPLTGAGRR1349 pKa = 11.84YY1350 pKa = 8.87LNQATVDD1357 pKa = 3.67AQGVLSGQTSASNPQLTDD1375 pKa = 3.33LSDD1378 pKa = 3.49NGTSADD1384 pKa = 3.44ANGNGRR1390 pKa = 11.84GNEE1393 pKa = 3.97AGEE1396 pKa = 4.14NDD1398 pKa = 4.35PTPVTPVYY1406 pKa = 10.76NPAITLTKK1414 pKa = 10.58AIDD1417 pKa = 3.93LSVLPSPIAADD1428 pKa = 3.62SPISFTFTVTNSGNITLTDD1447 pKa = 3.47VTLSDD1452 pKa = 3.81ALAGAVVTGGPITLAPGQVDD1472 pKa = 3.73STSFTATYY1480 pKa = 9.24PLSAADD1486 pKa = 3.68LTANTLSNTATATGTWGLDD1505 pKa = 3.16AGGNPQKK1512 pKa = 10.96VSDD1515 pKa = 4.77PDD1517 pKa = 3.68TVATSFADD1525 pKa = 3.21IALVKK1530 pKa = 10.55AADD1533 pKa = 3.68ASLLSVPPQVGDD1545 pKa = 3.6IISYY1549 pKa = 10.83DD1550 pKa = 3.53FAVTNTGNVPLTNVTLSDD1568 pKa = 3.43ILTDD1572 pKa = 3.59AVLTGGPIPSLAVGATDD1589 pKa = 3.8SSTFTATYY1597 pKa = 10.97ALTQADD1603 pKa = 3.42IDD1605 pKa = 3.77AGEE1608 pKa = 4.36VINRR1612 pKa = 11.84ATADD1616 pKa = 3.56GVYY1619 pKa = 9.34GTDD1622 pKa = 4.69DD1623 pKa = 3.9GGDD1626 pKa = 3.95PISVQDD1632 pKa = 3.54EE1633 pKa = 4.16SGANVTEE1640 pKa = 4.73DD1641 pKa = 3.48ADD1643 pKa = 4.19TVVPLTQAPRR1653 pKa = 11.84IEE1655 pKa = 4.85LIKK1658 pKa = 10.79SLSSLVDD1665 pKa = 3.27TTGDD1669 pKa = 3.68GQIGAGDD1676 pKa = 3.52TANYY1680 pKa = 9.57GFTVTNTGNVALAGVTVSDD1699 pKa = 4.34LLVSVSGGPVSLAIGATDD1717 pKa = 3.43STSFTAAYY1725 pKa = 10.45VLTQADD1731 pKa = 3.37VDD1733 pKa = 3.69RR1734 pKa = 11.84GYY1736 pKa = 10.97VQNTATATGAAVTSSGDD1753 pKa = 3.79PILDD1757 pKa = 3.35
Molecular weight: 174.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C8S4X4|C8S4X4_9RHOB Short-chain dehydrogenase/reductase SDR OS=Rhodobacter sp. SW2 OX=371731 GN=Rsw2DRAFT_3102 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.34GGRR28 pKa = 11.84LVLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 8.83KK41 pKa = 10.63LSAA44 pKa = 3.95
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.34GGRR28 pKa = 11.84LVLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 8.83KK41 pKa = 10.63LSAA44 pKa = 3.95
Molecular weight: 5.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1062988 |
30 |
2893 |
312.2 |
33.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.909 ± 0.075 | 0.897 ± 0.014 |
5.673 ± 0.042 | 5.093 ± 0.036 |
3.576 ± 0.029 | 9.009 ± 0.06 |
1.994 ± 0.022 | 4.74 ± 0.03 |
2.869 ± 0.032 | 10.72 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.652 ± 0.019 | 2.4 ± 0.026 |
5.346 ± 0.037 | 3.262 ± 0.023 |
6.803 ± 0.049 | 4.768 ± 0.031 |
5.358 ± 0.039 | 7.48 ± 0.035 |
1.414 ± 0.017 | 2.038 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
