
Capsella rubella
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
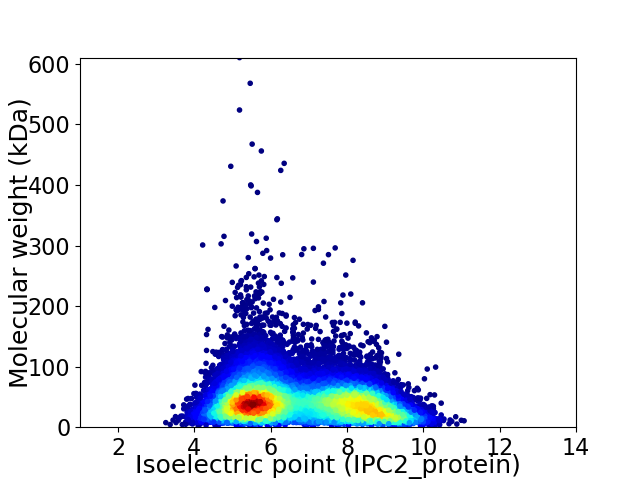
Virtual 2D-PAGE plot for 28039 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R0GIK7|R0GIK7_9BRAS Uncharacterized protein OS=Capsella rubella OX=81985 GN=CARUB_v10020743mg PE=4 SV=1
MM1 pKa = 7.56AGDD4 pKa = 4.11EE5 pKa = 4.43PKK7 pKa = 10.81SSAVKK12 pKa = 10.51DD13 pKa = 4.12DD14 pKa = 4.3ADD16 pKa = 3.83EE17 pKa = 4.93DD18 pKa = 4.95PYY20 pKa = 11.44NHH22 pKa = 7.5PYY24 pKa = 10.98DD25 pKa = 3.87GFGWFQNVMDD35 pKa = 5.32DD36 pKa = 4.14PNPNLMDD43 pKa = 3.41MAFTGNTSPSMCPPLLPLDD62 pKa = 3.8VDD64 pKa = 3.76QTQLFHH70 pKa = 7.15EE71 pKa = 5.22LDD73 pKa = 3.9LSISNYY79 pKa = 8.24DD80 pKa = 3.54TNDD83 pKa = 3.41DD84 pKa = 3.84FDD86 pKa = 4.03VLSFFDD92 pKa = 5.41DD93 pKa = 5.4GITTSLDD100 pKa = 3.12MLEE103 pKa = 4.76PLNDD107 pKa = 3.94GPNEE111 pKa = 4.01NVNNDD116 pKa = 3.25SQGEE120 pKa = 4.02ITYY123 pKa = 10.39DD124 pKa = 3.36YY125 pKa = 11.87VNDD128 pKa = 3.94HH129 pKa = 6.56NNNNVGEE136 pKa = 4.39TTRR139 pKa = 11.84LEE141 pKa = 3.83VSTRR145 pKa = 11.84VFGDD149 pKa = 3.41PNGPNYY155 pKa = 10.3EE156 pKa = 4.06IGGTSTTPVASDD168 pKa = 3.42KK169 pKa = 11.5DD170 pKa = 4.14FACDD174 pKa = 3.16CCTMLRR180 pKa = 11.84EE181 pKa = 4.09LVHH184 pKa = 5.8MKK186 pKa = 9.98EE187 pKa = 4.17GEE189 pKa = 3.99IMTTLVIYY197 pKa = 10.31GGIGFICHH205 pKa = 6.81AFLHH209 pKa = 5.92TRR211 pKa = 11.84LLPSDD216 pKa = 3.62STEE219 pKa = 4.02PQPQNQPEE227 pKa = 4.15PQKK230 pKa = 10.59IHH232 pKa = 7.41LMDD235 pKa = 3.73LTMEE239 pKa = 4.16EE240 pKa = 4.07VKK242 pKa = 10.79KK243 pKa = 10.8FIEE246 pKa = 5.11DD247 pKa = 3.61YY248 pKa = 10.65CSQRR252 pKa = 11.84LASGLSLVQDD262 pKa = 3.98TNAAFYY268 pKa = 10.73EE269 pKa = 4.25AMSVNSIFNQSPPMLAPSTFTEE291 pKa = 4.2ALDD294 pKa = 3.73VSLVPLNVGLATEE307 pKa = 4.55EE308 pKa = 4.1PTAA311 pKa = 4.21
MM1 pKa = 7.56AGDD4 pKa = 4.11EE5 pKa = 4.43PKK7 pKa = 10.81SSAVKK12 pKa = 10.51DD13 pKa = 4.12DD14 pKa = 4.3ADD16 pKa = 3.83EE17 pKa = 4.93DD18 pKa = 4.95PYY20 pKa = 11.44NHH22 pKa = 7.5PYY24 pKa = 10.98DD25 pKa = 3.87GFGWFQNVMDD35 pKa = 5.32DD36 pKa = 4.14PNPNLMDD43 pKa = 3.41MAFTGNTSPSMCPPLLPLDD62 pKa = 3.8VDD64 pKa = 3.76QTQLFHH70 pKa = 7.15EE71 pKa = 5.22LDD73 pKa = 3.9LSISNYY79 pKa = 8.24DD80 pKa = 3.54TNDD83 pKa = 3.41DD84 pKa = 3.84FDD86 pKa = 4.03VLSFFDD92 pKa = 5.41DD93 pKa = 5.4GITTSLDD100 pKa = 3.12MLEE103 pKa = 4.76PLNDD107 pKa = 3.94GPNEE111 pKa = 4.01NVNNDD116 pKa = 3.25SQGEE120 pKa = 4.02ITYY123 pKa = 10.39DD124 pKa = 3.36YY125 pKa = 11.87VNDD128 pKa = 3.94HH129 pKa = 6.56NNNNVGEE136 pKa = 4.39TTRR139 pKa = 11.84LEE141 pKa = 3.83VSTRR145 pKa = 11.84VFGDD149 pKa = 3.41PNGPNYY155 pKa = 10.3EE156 pKa = 4.06IGGTSTTPVASDD168 pKa = 3.42KK169 pKa = 11.5DD170 pKa = 4.14FACDD174 pKa = 3.16CCTMLRR180 pKa = 11.84EE181 pKa = 4.09LVHH184 pKa = 5.8MKK186 pKa = 9.98EE187 pKa = 4.17GEE189 pKa = 3.99IMTTLVIYY197 pKa = 10.31GGIGFICHH205 pKa = 6.81AFLHH209 pKa = 5.92TRR211 pKa = 11.84LLPSDD216 pKa = 3.62STEE219 pKa = 4.02PQPQNQPEE227 pKa = 4.15PQKK230 pKa = 10.59IHH232 pKa = 7.41LMDD235 pKa = 3.73LTMEE239 pKa = 4.16EE240 pKa = 4.07VKK242 pKa = 10.79KK243 pKa = 10.8FIEE246 pKa = 5.11DD247 pKa = 3.61YY248 pKa = 10.65CSQRR252 pKa = 11.84LASGLSLVQDD262 pKa = 3.98TNAAFYY268 pKa = 10.73EE269 pKa = 4.25AMSVNSIFNQSPPMLAPSTFTEE291 pKa = 4.2ALDD294 pKa = 3.73VSLVPLNVGLATEE307 pKa = 4.55EE308 pKa = 4.1PTAA311 pKa = 4.21
Molecular weight: 34.35 kDa
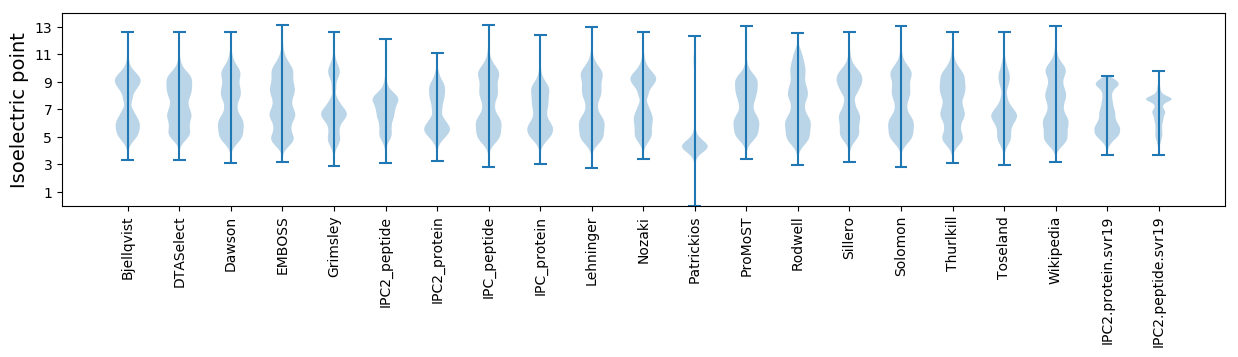
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R0IG64|R0IG64_9BRAS HVA22-like protein OS=Capsella rubella OX=81985 GN=CARUB_v10021029mg PE=3 SV=1
FF1 pKa = 6.56THH3 pKa = 7.1LHH5 pKa = 4.71TTHH8 pKa = 7.17TFTNLRR14 pKa = 11.84LLLTTRR20 pKa = 11.84LLNRR24 pKa = 11.84LLKK27 pKa = 10.61RR28 pKa = 11.84SLTTIQKK35 pKa = 7.75KK36 pKa = 7.34TKK38 pKa = 9.41NRR40 pKa = 11.84LVII43 pKa = 3.97
FF1 pKa = 6.56THH3 pKa = 7.1LHH5 pKa = 4.71TTHH8 pKa = 7.17TFTNLRR14 pKa = 11.84LLLTTRR20 pKa = 11.84LLNRR24 pKa = 11.84LLKK27 pKa = 10.61RR28 pKa = 11.84SLTTIQKK35 pKa = 7.75KK36 pKa = 7.34TKK38 pKa = 9.41NRR40 pKa = 11.84LVII43 pKa = 3.97
Molecular weight: 5.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
11560300 |
8 |
5389 |
412.3 |
46.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.323 ± 0.013 | 1.879 ± 0.008 |
5.408 ± 0.011 | 6.686 ± 0.019 |
4.275 ± 0.01 | 6.352 ± 0.012 |
2.291 ± 0.005 | 5.272 ± 0.011 |
6.373 ± 0.014 | 9.578 ± 0.017 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.45 ± 0.006 | 4.387 ± 0.01 |
4.802 ± 0.013 | 3.482 ± 0.01 |
5.404 ± 0.011 | 9.159 ± 0.018 |
5.106 ± 0.009 | 6.689 ± 0.01 |
1.25 ± 0.005 | 2.835 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
