
Hubei sobemo-like virus 33
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
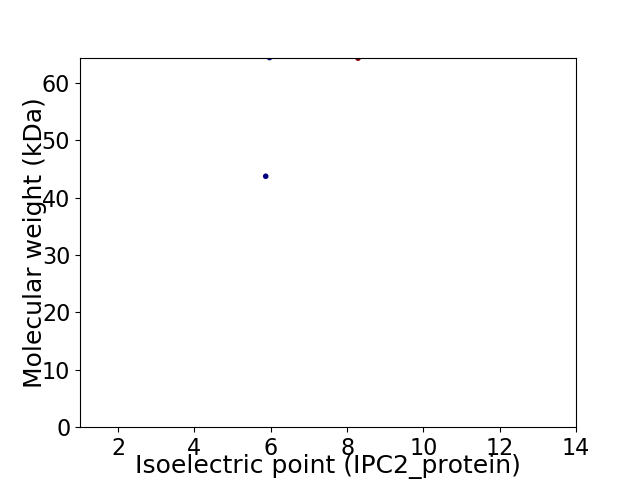
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
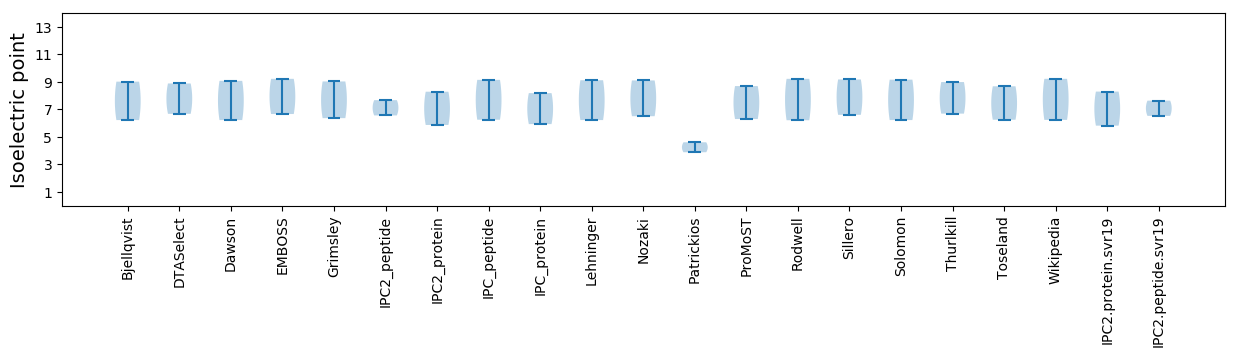
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KEF7|A0A1L3KEF7_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 33 OX=1923220 PE=4 SV=1
MM1 pKa = 7.51PGKK4 pKa = 9.42MDD6 pKa = 5.8LIHH9 pKa = 8.25IEE11 pKa = 4.42EE12 pKa = 4.27ILLNRR17 pKa = 11.84WKK19 pKa = 10.45AARR22 pKa = 11.84WRR24 pKa = 11.84IPDD27 pKa = 3.83DD28 pKa = 3.57FLEE31 pKa = 4.52RR32 pKa = 11.84THH34 pKa = 6.67FEE36 pKa = 4.12RR37 pKa = 11.84VVLEE41 pKa = 4.04IDD43 pKa = 3.3WTSSPGYY50 pKa = 9.28PWLLQHH56 pKa = 5.92TTNSAFFEE64 pKa = 4.52VKK66 pKa = 10.16DD67 pKa = 4.06GKK69 pKa = 9.78PSKK72 pKa = 10.5AALDD76 pKa = 3.92RR77 pKa = 11.84VWTIVQQRR85 pKa = 11.84LQSRR89 pKa = 11.84EE90 pKa = 3.93CDD92 pKa = 4.03PIRR95 pKa = 11.84LFIKK99 pKa = 10.28PEE101 pKa = 3.49PHH103 pKa = 6.05KK104 pKa = 10.66QKK106 pKa = 11.03KK107 pKa = 9.65LDD109 pKa = 3.53SKK111 pKa = 11.1AYY113 pKa = 9.65RR114 pKa = 11.84LISSVSVIDD123 pKa = 4.07QIIDD127 pKa = 3.25ALLFGEE133 pKa = 4.6MNQRR137 pKa = 11.84MIEE140 pKa = 4.08RR141 pKa = 11.84YY142 pKa = 9.69LDD144 pKa = 3.47VPGKK148 pKa = 10.38VGWSPYY154 pKa = 8.79VGGWKK159 pKa = 9.98IMPAYY164 pKa = 10.51GNVSLDD170 pKa = 3.05KK171 pKa = 11.14SAWDD175 pKa = 3.32WSVNAWIIQCILNMRR190 pKa = 11.84MQLCDD195 pKa = 3.49NVSEE199 pKa = 4.23SWIDD203 pKa = 3.51LACWRR208 pKa = 11.84YY209 pKa = 8.74NALYY213 pKa = 10.16NSPLFVTSGGLLLRR227 pKa = 11.84QLQPGVVKK235 pKa = 10.42SGCYY239 pKa = 7.84NTIADD244 pKa = 3.88NSIAQDD250 pKa = 3.77LLHH253 pKa = 6.72VRR255 pKa = 11.84VCVEE259 pKa = 4.29LDD261 pKa = 3.5IEE263 pKa = 4.88CGDD266 pKa = 4.21LMSMGDD272 pKa = 3.88DD273 pKa = 3.32TTQRR277 pKa = 11.84WFSEE281 pKa = 4.29FPDD284 pKa = 3.64YY285 pKa = 11.33VEE287 pKa = 5.78RR288 pKa = 11.84LSQYY292 pKa = 9.62CHH294 pKa = 5.8VKK296 pKa = 10.5GSVNGTEE303 pKa = 4.31FAGHH307 pKa = 7.27RR308 pKa = 11.84FTSCSVEE315 pKa = 3.63PLYY318 pKa = 10.72KK319 pKa = 10.42GKK321 pKa = 9.56HH322 pKa = 5.99AYY324 pKa = 10.71NIMNMNPKK332 pKa = 10.14YY333 pKa = 10.68GEE335 pKa = 3.93EE336 pKa = 4.04TAASYY341 pKa = 11.51ALLYY345 pKa = 9.92HH346 pKa = 6.8RR347 pKa = 11.84SVHH350 pKa = 5.74SNYY353 pKa = 9.07IRR355 pKa = 11.84ALLEE359 pKa = 3.88RR360 pKa = 11.84MGYY363 pKa = 7.08TLPSRR368 pKa = 11.84QTLDD372 pKa = 3.22VIYY375 pKa = 10.62DD376 pKa = 3.83GEE378 pKa = 4.36VV379 pKa = 2.61
MM1 pKa = 7.51PGKK4 pKa = 9.42MDD6 pKa = 5.8LIHH9 pKa = 8.25IEE11 pKa = 4.42EE12 pKa = 4.27ILLNRR17 pKa = 11.84WKK19 pKa = 10.45AARR22 pKa = 11.84WRR24 pKa = 11.84IPDD27 pKa = 3.83DD28 pKa = 3.57FLEE31 pKa = 4.52RR32 pKa = 11.84THH34 pKa = 6.67FEE36 pKa = 4.12RR37 pKa = 11.84VVLEE41 pKa = 4.04IDD43 pKa = 3.3WTSSPGYY50 pKa = 9.28PWLLQHH56 pKa = 5.92TTNSAFFEE64 pKa = 4.52VKK66 pKa = 10.16DD67 pKa = 4.06GKK69 pKa = 9.78PSKK72 pKa = 10.5AALDD76 pKa = 3.92RR77 pKa = 11.84VWTIVQQRR85 pKa = 11.84LQSRR89 pKa = 11.84EE90 pKa = 3.93CDD92 pKa = 4.03PIRR95 pKa = 11.84LFIKK99 pKa = 10.28PEE101 pKa = 3.49PHH103 pKa = 6.05KK104 pKa = 10.66QKK106 pKa = 11.03KK107 pKa = 9.65LDD109 pKa = 3.53SKK111 pKa = 11.1AYY113 pKa = 9.65RR114 pKa = 11.84LISSVSVIDD123 pKa = 4.07QIIDD127 pKa = 3.25ALLFGEE133 pKa = 4.6MNQRR137 pKa = 11.84MIEE140 pKa = 4.08RR141 pKa = 11.84YY142 pKa = 9.69LDD144 pKa = 3.47VPGKK148 pKa = 10.38VGWSPYY154 pKa = 8.79VGGWKK159 pKa = 9.98IMPAYY164 pKa = 10.51GNVSLDD170 pKa = 3.05KK171 pKa = 11.14SAWDD175 pKa = 3.32WSVNAWIIQCILNMRR190 pKa = 11.84MQLCDD195 pKa = 3.49NVSEE199 pKa = 4.23SWIDD203 pKa = 3.51LACWRR208 pKa = 11.84YY209 pKa = 8.74NALYY213 pKa = 10.16NSPLFVTSGGLLLRR227 pKa = 11.84QLQPGVVKK235 pKa = 10.42SGCYY239 pKa = 7.84NTIADD244 pKa = 3.88NSIAQDD250 pKa = 3.77LLHH253 pKa = 6.72VRR255 pKa = 11.84VCVEE259 pKa = 4.29LDD261 pKa = 3.5IEE263 pKa = 4.88CGDD266 pKa = 4.21LMSMGDD272 pKa = 3.88DD273 pKa = 3.32TTQRR277 pKa = 11.84WFSEE281 pKa = 4.29FPDD284 pKa = 3.64YY285 pKa = 11.33VEE287 pKa = 5.78RR288 pKa = 11.84LSQYY292 pKa = 9.62CHH294 pKa = 5.8VKK296 pKa = 10.5GSVNGTEE303 pKa = 4.31FAGHH307 pKa = 7.27RR308 pKa = 11.84FTSCSVEE315 pKa = 3.63PLYY318 pKa = 10.72KK319 pKa = 10.42GKK321 pKa = 9.56HH322 pKa = 5.99AYY324 pKa = 10.71NIMNMNPKK332 pKa = 10.14YY333 pKa = 10.68GEE335 pKa = 3.93EE336 pKa = 4.04TAASYY341 pKa = 11.51ALLYY345 pKa = 9.92HH346 pKa = 6.8RR347 pKa = 11.84SVHH350 pKa = 5.74SNYY353 pKa = 9.07IRR355 pKa = 11.84ALLEE359 pKa = 3.88RR360 pKa = 11.84MGYY363 pKa = 7.08TLPSRR368 pKa = 11.84QTLDD372 pKa = 3.22VIYY375 pKa = 10.62DD376 pKa = 3.83GEE378 pKa = 4.36VV379 pKa = 2.61
Molecular weight: 43.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEF7|A0A1L3KEF7_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 33 OX=1923220 PE=4 SV=1
MM1 pKa = 7.36FPLIAMIKK9 pKa = 8.71VFCCVLTAGWLRR21 pKa = 11.84YY22 pKa = 9.11IYY24 pKa = 10.35LRR26 pKa = 11.84VEE28 pKa = 4.57MIPGLLAAWLIRR40 pKa = 11.84MEE42 pKa = 4.75EE43 pKa = 3.8KK44 pKa = 10.48WMDD47 pKa = 3.95ALDD50 pKa = 3.49AWVCRR55 pKa = 11.84IALGPSEE62 pKa = 4.99KK63 pKa = 9.88PAPVPLTFWEE73 pKa = 4.31EE74 pKa = 3.75MVYY77 pKa = 9.64TLSPYY82 pKa = 9.98FDD84 pKa = 3.26VAIRR88 pKa = 11.84RR89 pKa = 11.84LVFIGYY95 pKa = 9.23SLTAIFVVVLAFMAVKK111 pKa = 10.23KK112 pKa = 9.8PLRR115 pKa = 11.84RR116 pKa = 11.84TVLRR120 pKa = 11.84LRR122 pKa = 11.84GITFEE127 pKa = 4.02SMQPGSEE134 pKa = 4.14FVEE137 pKa = 4.81GAVPDD142 pKa = 4.37FQVGIYY148 pKa = 10.27DD149 pKa = 3.75AGTFVDD155 pKa = 3.75TFIGYY160 pKa = 9.06GIRR163 pKa = 11.84FGSLLVLPAHH173 pKa = 6.43VIKK176 pKa = 10.51HH177 pKa = 5.04VKK179 pKa = 9.38QLVIEE184 pKa = 4.47GRR186 pKa = 11.84TGRR189 pKa = 11.84LGLNTSYY196 pKa = 10.72IPSKK200 pKa = 10.87VSTDD204 pKa = 3.28LVYY207 pKa = 10.62IPMTEE212 pKa = 4.19AQWSRR217 pKa = 11.84VGTSTAKK224 pKa = 10.27VPKK227 pKa = 9.99KK228 pKa = 10.28LVNSLVSCTGRR239 pKa = 11.84KK240 pKa = 7.59GTSTGLLTQTDD251 pKa = 3.86VLGIMKK257 pKa = 10.33YY258 pKa = 10.14SGSTISGMSGAAYY271 pKa = 10.43YY272 pKa = 10.5SGQTVYY278 pKa = 11.36GMHH281 pKa = 6.62TGVAGDD287 pKa = 3.67YY288 pKa = 10.6NIGVTATLIMAEE300 pKa = 4.25TRR302 pKa = 11.84RR303 pKa = 11.84LVLGEE308 pKa = 4.44SPTLDD313 pKa = 2.98EE314 pKa = 4.36MKK316 pKa = 10.74GLNRR320 pKa = 11.84ASAAKK325 pKa = 9.93VRR327 pKa = 11.84SGWDD331 pKa = 3.13TTHH334 pKa = 7.21LMKK337 pKa = 10.73QVDD340 pKa = 3.71SMYY343 pKa = 10.94ADD345 pKa = 4.33EE346 pKa = 5.36IGWAADD352 pKa = 3.19EE353 pKa = 4.16EE354 pKa = 4.35MDD356 pKa = 4.78YY357 pKa = 11.36GVQLTFDD364 pKa = 4.12GEE366 pKa = 4.18ASEE369 pKa = 4.19EE370 pKa = 4.26AIEE373 pKa = 3.91KK374 pKa = 10.09WMTFFTAMPSMQRR387 pKa = 11.84EE388 pKa = 4.03SAINLLQSYY397 pKa = 8.8TNASRR402 pKa = 11.84AAKK405 pKa = 9.48GQSDD409 pKa = 4.67EE410 pKa = 4.43PTPITLPKK418 pKa = 10.37DD419 pKa = 3.87FVSMRR424 pKa = 11.84LDD426 pKa = 4.08AIEE429 pKa = 4.45KK430 pKa = 10.34RR431 pKa = 11.84LDD433 pKa = 3.74KK434 pKa = 10.23ITALEE439 pKa = 3.97QRR441 pKa = 11.84VKK443 pKa = 10.03TLEE446 pKa = 3.93EE447 pKa = 4.37KK448 pKa = 10.39IVAGVRR454 pKa = 11.84KK455 pKa = 9.34PGIAIPNHH463 pKa = 5.36SPKK466 pKa = 10.22PFPCGYY472 pKa = 8.37EE473 pKa = 3.68KK474 pKa = 10.64CYY476 pKa = 10.66KK477 pKa = 10.56AFNKK481 pKa = 10.13PEE483 pKa = 3.62ARR485 pKa = 11.84MAHH488 pKa = 5.01QVAVGHH494 pKa = 5.51VVGEE498 pKa = 4.41SAFGADD504 pKa = 3.92EE505 pKa = 4.39KK506 pKa = 10.26PTVGTAPSPVFRR518 pKa = 11.84QRR520 pKa = 11.84PLQKK524 pKa = 10.01KK525 pKa = 9.55KK526 pKa = 10.4PSTSSRR532 pKa = 11.84NSSSSQEE539 pKa = 3.7RR540 pKa = 11.84GKK542 pKa = 10.03PSTSQQQNQSDD553 pKa = 4.13TLVSLMRR560 pKa = 11.84TIGDD564 pKa = 3.63LQKK567 pKa = 11.11SLEE570 pKa = 4.19ATVGLLSATTQNSTVV585 pKa = 3.11
MM1 pKa = 7.36FPLIAMIKK9 pKa = 8.71VFCCVLTAGWLRR21 pKa = 11.84YY22 pKa = 9.11IYY24 pKa = 10.35LRR26 pKa = 11.84VEE28 pKa = 4.57MIPGLLAAWLIRR40 pKa = 11.84MEE42 pKa = 4.75EE43 pKa = 3.8KK44 pKa = 10.48WMDD47 pKa = 3.95ALDD50 pKa = 3.49AWVCRR55 pKa = 11.84IALGPSEE62 pKa = 4.99KK63 pKa = 9.88PAPVPLTFWEE73 pKa = 4.31EE74 pKa = 3.75MVYY77 pKa = 9.64TLSPYY82 pKa = 9.98FDD84 pKa = 3.26VAIRR88 pKa = 11.84RR89 pKa = 11.84LVFIGYY95 pKa = 9.23SLTAIFVVVLAFMAVKK111 pKa = 10.23KK112 pKa = 9.8PLRR115 pKa = 11.84RR116 pKa = 11.84TVLRR120 pKa = 11.84LRR122 pKa = 11.84GITFEE127 pKa = 4.02SMQPGSEE134 pKa = 4.14FVEE137 pKa = 4.81GAVPDD142 pKa = 4.37FQVGIYY148 pKa = 10.27DD149 pKa = 3.75AGTFVDD155 pKa = 3.75TFIGYY160 pKa = 9.06GIRR163 pKa = 11.84FGSLLVLPAHH173 pKa = 6.43VIKK176 pKa = 10.51HH177 pKa = 5.04VKK179 pKa = 9.38QLVIEE184 pKa = 4.47GRR186 pKa = 11.84TGRR189 pKa = 11.84LGLNTSYY196 pKa = 10.72IPSKK200 pKa = 10.87VSTDD204 pKa = 3.28LVYY207 pKa = 10.62IPMTEE212 pKa = 4.19AQWSRR217 pKa = 11.84VGTSTAKK224 pKa = 10.27VPKK227 pKa = 9.99KK228 pKa = 10.28LVNSLVSCTGRR239 pKa = 11.84KK240 pKa = 7.59GTSTGLLTQTDD251 pKa = 3.86VLGIMKK257 pKa = 10.33YY258 pKa = 10.14SGSTISGMSGAAYY271 pKa = 10.43YY272 pKa = 10.5SGQTVYY278 pKa = 11.36GMHH281 pKa = 6.62TGVAGDD287 pKa = 3.67YY288 pKa = 10.6NIGVTATLIMAEE300 pKa = 4.25TRR302 pKa = 11.84RR303 pKa = 11.84LVLGEE308 pKa = 4.44SPTLDD313 pKa = 2.98EE314 pKa = 4.36MKK316 pKa = 10.74GLNRR320 pKa = 11.84ASAAKK325 pKa = 9.93VRR327 pKa = 11.84SGWDD331 pKa = 3.13TTHH334 pKa = 7.21LMKK337 pKa = 10.73QVDD340 pKa = 3.71SMYY343 pKa = 10.94ADD345 pKa = 4.33EE346 pKa = 5.36IGWAADD352 pKa = 3.19EE353 pKa = 4.16EE354 pKa = 4.35MDD356 pKa = 4.78YY357 pKa = 11.36GVQLTFDD364 pKa = 4.12GEE366 pKa = 4.18ASEE369 pKa = 4.19EE370 pKa = 4.26AIEE373 pKa = 3.91KK374 pKa = 10.09WMTFFTAMPSMQRR387 pKa = 11.84EE388 pKa = 4.03SAINLLQSYY397 pKa = 8.8TNASRR402 pKa = 11.84AAKK405 pKa = 9.48GQSDD409 pKa = 4.67EE410 pKa = 4.43PTPITLPKK418 pKa = 10.37DD419 pKa = 3.87FVSMRR424 pKa = 11.84LDD426 pKa = 4.08AIEE429 pKa = 4.45KK430 pKa = 10.34RR431 pKa = 11.84LDD433 pKa = 3.74KK434 pKa = 10.23ITALEE439 pKa = 3.97QRR441 pKa = 11.84VKK443 pKa = 10.03TLEE446 pKa = 3.93EE447 pKa = 4.37KK448 pKa = 10.39IVAGVRR454 pKa = 11.84KK455 pKa = 9.34PGIAIPNHH463 pKa = 5.36SPKK466 pKa = 10.22PFPCGYY472 pKa = 8.37EE473 pKa = 3.68KK474 pKa = 10.64CYY476 pKa = 10.66KK477 pKa = 10.56AFNKK481 pKa = 10.13PEE483 pKa = 3.62ARR485 pKa = 11.84MAHH488 pKa = 5.01QVAVGHH494 pKa = 5.51VVGEE498 pKa = 4.41SAFGADD504 pKa = 3.92EE505 pKa = 4.39KK506 pKa = 10.26PTVGTAPSPVFRR518 pKa = 11.84QRR520 pKa = 11.84PLQKK524 pKa = 10.01KK525 pKa = 9.55KK526 pKa = 10.4PSTSSRR532 pKa = 11.84NSSSSQEE539 pKa = 3.7RR540 pKa = 11.84GKK542 pKa = 10.03PSTSQQQNQSDD553 pKa = 4.13TLVSLMRR560 pKa = 11.84TIGDD564 pKa = 3.63LQKK567 pKa = 11.11SLEE570 pKa = 4.19ATVGLLSATTQNSTVV585 pKa = 3.11
Molecular weight: 64.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
964 |
379 |
585 |
482.0 |
54.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.054 ± 1.071 | 1.556 ± 0.493 |
4.876 ± 0.878 | 5.602 ± 0.037 |
3.32 ± 0.251 | 6.95 ± 0.69 |
1.763 ± 0.527 | 5.705 ± 0.378 |
5.394 ± 0.389 | 9.025 ± 0.445 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.631 ± 0.28 | 2.801 ± 0.856 |
4.979 ± 0.298 | 3.838 ± 0.072 |
5.602 ± 0.281 | 7.884 ± 0.14 |
6.224 ± 1.525 | 7.676 ± 0.492 |
2.282 ± 0.692 | 3.838 ± 0.549 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
