
Beihai rhabdo-like virus 6
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Nyamiviridae; Crustavirus; Beihai crustavirus
Average proteome isoelectric point is 7.19
Get precalculated fractions of proteins
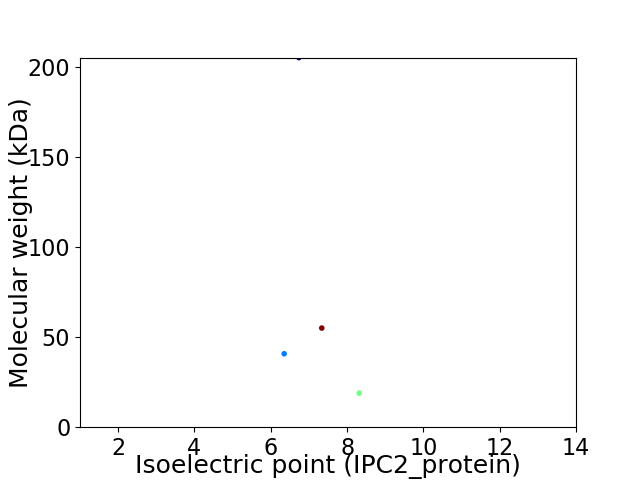
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KMN0|A0A1L3KMN0_9MONO Putative glycoprotein OS=Beihai rhabdo-like virus 6 OX=1922656 PE=4 SV=1
MM1 pKa = 6.85TAAGPFTKK9 pKa = 10.13RR10 pKa = 11.84PRR12 pKa = 11.84HH13 pKa = 5.43YY14 pKa = 10.15EE15 pKa = 3.6SDD17 pKa = 3.41MVSLKK22 pKa = 10.36RR23 pKa = 11.84HH24 pKa = 5.77APSFTPLQMPHH35 pKa = 6.92EE36 pKa = 4.37IDD38 pKa = 3.15IRR40 pKa = 11.84SRR42 pKa = 11.84LYY44 pKa = 11.04NGTTTAIDD52 pKa = 3.18WVAAVAYY59 pKa = 8.77QYY61 pKa = 11.09RR62 pKa = 11.84LSSEE66 pKa = 4.42VIAKK70 pKa = 9.58LFSPYY75 pKa = 11.13ALDD78 pKa = 3.26QHH80 pKa = 6.34NRR82 pKa = 11.84PLYY85 pKa = 9.9FPRR88 pKa = 11.84FDD90 pKa = 3.73TEE92 pKa = 3.86DD93 pKa = 3.25AKK95 pKa = 10.83IANIRR100 pKa = 11.84ARR102 pKa = 11.84VKK104 pKa = 10.93DD105 pKa = 3.5SGKK108 pKa = 10.64EE109 pKa = 3.74GLEE112 pKa = 3.79AARR115 pKa = 11.84VGIQIAAYY123 pKa = 8.64AVIVGLHH130 pKa = 5.25KK131 pKa = 9.74TIEE134 pKa = 4.53DD135 pKa = 3.6KK136 pKa = 11.32NKK138 pKa = 9.74EE139 pKa = 4.04YY140 pKa = 10.81VLRR143 pKa = 11.84RR144 pKa = 11.84IAAVGASQDD153 pKa = 4.76DD154 pKa = 4.45EE155 pKa = 4.51ILTNIPVADD164 pKa = 3.39MDD166 pKa = 3.77YY167 pKa = 10.02WVVVDD172 pKa = 6.16RR173 pKa = 11.84INDD176 pKa = 3.13AHH178 pKa = 7.33KK179 pKa = 10.22YY180 pKa = 8.89IRR182 pKa = 11.84AAEE185 pKa = 3.95ITNDD189 pKa = 4.0LLHH192 pKa = 6.95DD193 pKa = 3.85VCTSVDD199 pKa = 3.74DD200 pKa = 5.23DD201 pKa = 4.08STPLHH206 pKa = 6.54IAMVDD211 pKa = 3.74SIKK214 pKa = 10.3MVLKK218 pKa = 10.47GYY220 pKa = 10.97GMLGTQVMANFLTTPSKK237 pKa = 10.63AWLMAQVLDD246 pKa = 3.9EE247 pKa = 5.08GIAFKK252 pKa = 10.71RR253 pKa = 11.84ALRR256 pKa = 11.84TFKK259 pKa = 10.76ALHH262 pKa = 6.0GDD264 pKa = 2.81SWEE267 pKa = 3.87WGRR270 pKa = 11.84VLKK273 pKa = 11.08LNTIEE278 pKa = 4.39TLDD281 pKa = 3.6KK282 pKa = 10.48TSYY285 pKa = 9.59PHH287 pKa = 7.24LYY289 pKa = 7.91WAAAVRR295 pKa = 11.84RR296 pKa = 11.84NGGTGGQADD305 pKa = 4.04GLKK308 pKa = 10.05NYY310 pKa = 8.97VMKK313 pKa = 10.9APDD316 pKa = 3.46GLMVNKK322 pKa = 8.77TQLEE326 pKa = 4.22SWALKK331 pKa = 10.4DD332 pKa = 3.55LASAAVLTATFAQKK346 pKa = 10.14IKK348 pKa = 9.41EE349 pKa = 4.28TLGINLDD356 pKa = 3.95DD357 pKa = 4.05KK358 pKa = 11.77VGDD361 pKa = 3.84KK362 pKa = 11.42VEE364 pKa = 4.23DD365 pKa = 3.65LL366 pKa = 4.17
MM1 pKa = 6.85TAAGPFTKK9 pKa = 10.13RR10 pKa = 11.84PRR12 pKa = 11.84HH13 pKa = 5.43YY14 pKa = 10.15EE15 pKa = 3.6SDD17 pKa = 3.41MVSLKK22 pKa = 10.36RR23 pKa = 11.84HH24 pKa = 5.77APSFTPLQMPHH35 pKa = 6.92EE36 pKa = 4.37IDD38 pKa = 3.15IRR40 pKa = 11.84SRR42 pKa = 11.84LYY44 pKa = 11.04NGTTTAIDD52 pKa = 3.18WVAAVAYY59 pKa = 8.77QYY61 pKa = 11.09RR62 pKa = 11.84LSSEE66 pKa = 4.42VIAKK70 pKa = 9.58LFSPYY75 pKa = 11.13ALDD78 pKa = 3.26QHH80 pKa = 6.34NRR82 pKa = 11.84PLYY85 pKa = 9.9FPRR88 pKa = 11.84FDD90 pKa = 3.73TEE92 pKa = 3.86DD93 pKa = 3.25AKK95 pKa = 10.83IANIRR100 pKa = 11.84ARR102 pKa = 11.84VKK104 pKa = 10.93DD105 pKa = 3.5SGKK108 pKa = 10.64EE109 pKa = 3.74GLEE112 pKa = 3.79AARR115 pKa = 11.84VGIQIAAYY123 pKa = 8.64AVIVGLHH130 pKa = 5.25KK131 pKa = 9.74TIEE134 pKa = 4.53DD135 pKa = 3.6KK136 pKa = 11.32NKK138 pKa = 9.74EE139 pKa = 4.04YY140 pKa = 10.81VLRR143 pKa = 11.84RR144 pKa = 11.84IAAVGASQDD153 pKa = 4.76DD154 pKa = 4.45EE155 pKa = 4.51ILTNIPVADD164 pKa = 3.39MDD166 pKa = 3.77YY167 pKa = 10.02WVVVDD172 pKa = 6.16RR173 pKa = 11.84INDD176 pKa = 3.13AHH178 pKa = 7.33KK179 pKa = 10.22YY180 pKa = 8.89IRR182 pKa = 11.84AAEE185 pKa = 3.95ITNDD189 pKa = 4.0LLHH192 pKa = 6.95DD193 pKa = 3.85VCTSVDD199 pKa = 3.74DD200 pKa = 5.23DD201 pKa = 4.08STPLHH206 pKa = 6.54IAMVDD211 pKa = 3.74SIKK214 pKa = 10.3MVLKK218 pKa = 10.47GYY220 pKa = 10.97GMLGTQVMANFLTTPSKK237 pKa = 10.63AWLMAQVLDD246 pKa = 3.9EE247 pKa = 5.08GIAFKK252 pKa = 10.71RR253 pKa = 11.84ALRR256 pKa = 11.84TFKK259 pKa = 10.76ALHH262 pKa = 6.0GDD264 pKa = 2.81SWEE267 pKa = 3.87WGRR270 pKa = 11.84VLKK273 pKa = 11.08LNTIEE278 pKa = 4.39TLDD281 pKa = 3.6KK282 pKa = 10.48TSYY285 pKa = 9.59PHH287 pKa = 7.24LYY289 pKa = 7.91WAAAVRR295 pKa = 11.84RR296 pKa = 11.84NGGTGGQADD305 pKa = 4.04GLKK308 pKa = 10.05NYY310 pKa = 8.97VMKK313 pKa = 10.9APDD316 pKa = 3.46GLMVNKK322 pKa = 8.77TQLEE326 pKa = 4.22SWALKK331 pKa = 10.4DD332 pKa = 3.55LASAAVLTATFAQKK346 pKa = 10.14IKK348 pKa = 9.41EE349 pKa = 4.28TLGINLDD356 pKa = 3.95DD357 pKa = 4.05KK358 pKa = 11.77VGDD361 pKa = 3.84KK362 pKa = 11.42VEE364 pKa = 4.23DD365 pKa = 3.65LL366 pKa = 4.17
Molecular weight: 40.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KMM4|A0A1L3KMM4_9MONO Putative nucleoprotein OS=Beihai rhabdo-like virus 6 OX=1922656 PE=4 SV=1
MM1 pKa = 7.58AEE3 pKa = 3.66KK4 pKa = 10.42SRR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.79KK9 pKa = 10.83SLGEE13 pKa = 3.88RR14 pKa = 11.84TPGSISARR22 pKa = 11.84VGDD25 pKa = 3.71RR26 pKa = 11.84AKK28 pKa = 10.51KK29 pKa = 9.83RR30 pKa = 11.84KK31 pKa = 8.3LTEE34 pKa = 4.13EE35 pKa = 4.2EE36 pKa = 4.63DD37 pKa = 3.85PLPAAIASFTEE48 pKa = 3.97LMEE51 pKa = 4.61GKK53 pKa = 9.74FARR56 pKa = 11.84VMEE59 pKa = 4.26ILEE62 pKa = 4.61EE63 pKa = 4.13IRR65 pKa = 11.84SQTAGMEE72 pKa = 4.0ARR74 pKa = 11.84ISSLEE79 pKa = 3.85RR80 pKa = 11.84RR81 pKa = 11.84VGNMNVDD88 pKa = 3.44PRR90 pKa = 11.84GTTVTAPPVATIGRR104 pKa = 11.84GRR106 pKa = 11.84GRR108 pKa = 11.84GLTLVPPRR116 pKa = 11.84QIGSTAAVGRR126 pKa = 11.84GRR128 pKa = 11.84GGRR131 pKa = 11.84GQPIAGPSSFRR142 pKa = 11.84PRR144 pKa = 11.84SPEE147 pKa = 3.68TGSDD151 pKa = 3.65DD152 pKa = 4.54EE153 pKa = 4.81EE154 pKa = 6.44DD155 pKa = 3.8GSAMDD160 pKa = 4.73GQPSDD165 pKa = 5.16DD166 pKa = 4.03RR167 pKa = 11.84YY168 pKa = 11.34EE169 pKa = 4.41DD170 pKa = 4.01ADD172 pKa = 4.66LLGLLL177 pKa = 4.64
MM1 pKa = 7.58AEE3 pKa = 3.66KK4 pKa = 10.42SRR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.79KK9 pKa = 10.83SLGEE13 pKa = 3.88RR14 pKa = 11.84TPGSISARR22 pKa = 11.84VGDD25 pKa = 3.71RR26 pKa = 11.84AKK28 pKa = 10.51KK29 pKa = 9.83RR30 pKa = 11.84KK31 pKa = 8.3LTEE34 pKa = 4.13EE35 pKa = 4.2EE36 pKa = 4.63DD37 pKa = 3.85PLPAAIASFTEE48 pKa = 3.97LMEE51 pKa = 4.61GKK53 pKa = 9.74FARR56 pKa = 11.84VMEE59 pKa = 4.26ILEE62 pKa = 4.61EE63 pKa = 4.13IRR65 pKa = 11.84SQTAGMEE72 pKa = 4.0ARR74 pKa = 11.84ISSLEE79 pKa = 3.85RR80 pKa = 11.84RR81 pKa = 11.84VGNMNVDD88 pKa = 3.44PRR90 pKa = 11.84GTTVTAPPVATIGRR104 pKa = 11.84GRR106 pKa = 11.84GRR108 pKa = 11.84GLTLVPPRR116 pKa = 11.84QIGSTAAVGRR126 pKa = 11.84GRR128 pKa = 11.84GGRR131 pKa = 11.84GQPIAGPSSFRR142 pKa = 11.84PRR144 pKa = 11.84SPEE147 pKa = 3.68TGSDD151 pKa = 3.65DD152 pKa = 4.54EE153 pKa = 4.81EE154 pKa = 6.44DD155 pKa = 3.8GSAMDD160 pKa = 4.73GQPSDD165 pKa = 5.16DD166 pKa = 4.03RR167 pKa = 11.84YY168 pKa = 11.34EE169 pKa = 4.41DD170 pKa = 4.01ADD172 pKa = 4.66LLGLLL177 pKa = 4.64
Molecular weight: 18.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2884 |
177 |
1832 |
721.0 |
80.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.079 ± 1.04 | 1.872 ± 0.619 |
5.34 ± 0.798 | 6.727 ± 0.727 |
3.467 ± 0.42 | 7.767 ± 1.32 |
2.739 ± 0.57 | 4.716 ± 0.627 |
5.617 ± 0.603 | 9.847 ± 0.544 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.462 ± 0.225 | 2.288 ± 0.337 |
4.993 ± 0.426 | 2.739 ± 0.336 |
6.657 ± 0.797 | 7.282 ± 0.634 |
5.895 ± 0.319 | 7.004 ± 0.387 |
1.63 ± 0.226 | 2.878 ± 0.448 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
