
Selenomonas sputigena (strain ATCC 35185 / DSM 20758 / VPI D19B-28)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Negativicutes; Selenomonadales; Selenomonadaceae; Selenomonas; Selenomonas sputigena
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
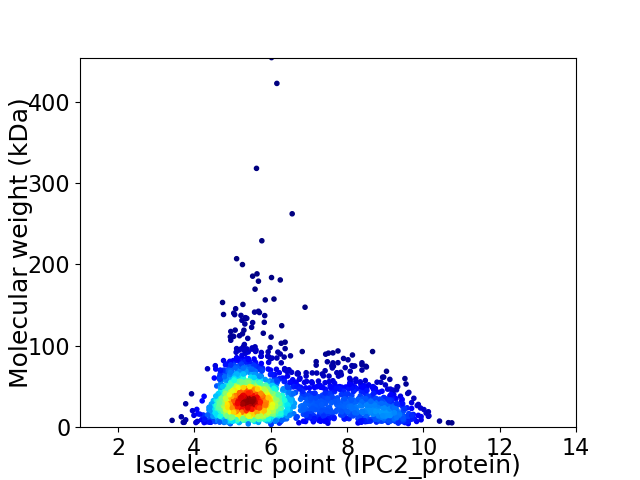
Virtual 2D-PAGE plot for 2243 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F4EWI3|F4EWI3_SELS3 Dynamin family protein OS=Selenomonas sputigena (strain ATCC 35185 / DSM 20758 / VPI D19B-28) OX=546271 GN=Selsp_2214 PE=4 SV=1
MM1 pKa = 7.51NRR3 pKa = 11.84FCRR6 pKa = 11.84ILRR9 pKa = 11.84QAFAVLLMTFIIAGGGYY26 pKa = 10.33GEE28 pKa = 4.28AVKK31 pKa = 10.86AGWCFAASLDD41 pKa = 4.45LAGLEE46 pKa = 4.38DD47 pKa = 3.94RR48 pKa = 11.84AVDD51 pKa = 3.8IDD53 pKa = 4.05PSDD56 pKa = 3.8GSVSVTLSEE65 pKa = 4.95SEE67 pKa = 4.22MEE69 pKa = 4.21LLSSVRR75 pKa = 11.84CNLLYY80 pKa = 10.59LDD82 pKa = 4.35EE83 pKa = 4.72NVVIHH88 pKa = 6.76LGSDD92 pKa = 3.31ANIDD96 pKa = 3.88ADD98 pKa = 3.21WDD100 pKa = 3.89TGVFRR105 pKa = 11.84DD106 pKa = 4.38NFDD109 pKa = 3.67GTWPMLDD116 pKa = 3.2GHH118 pKa = 6.32PVYY121 pKa = 10.52IEE123 pKa = 3.88LTEE126 pKa = 4.54EE127 pKa = 4.38GDD129 pKa = 3.99DD130 pKa = 3.76YY131 pKa = 11.75NLYY134 pKa = 10.6SIPIKK139 pKa = 10.96LNGEE143 pKa = 3.91EE144 pKa = 5.07CILQVSYY151 pKa = 11.25NYY153 pKa = 10.57NDD155 pKa = 3.36EE156 pKa = 4.44TYY158 pKa = 10.2HH159 pKa = 7.63ILGAQRR165 pKa = 11.84EE166 pKa = 4.32PEE168 pKa = 4.01EE169 pKa = 5.34SGMSDD174 pKa = 2.86QGLIPLRR181 pKa = 11.84EE182 pKa = 4.04GDD184 pKa = 3.99MVTTIHH190 pKa = 6.27FAKK193 pKa = 10.4SVSDD197 pKa = 3.85NDD199 pKa = 3.9DD200 pKa = 4.01DD201 pKa = 4.1EE202 pKa = 4.83LAAVEE207 pKa = 5.19AEE209 pKa = 4.12TFQIGSHH216 pKa = 6.32PAVEE220 pKa = 4.47DD221 pKa = 3.82EE222 pKa = 4.21EE223 pKa = 6.35LGDD226 pKa = 3.59GTYY229 pKa = 10.9GYY231 pKa = 9.91IFEE234 pKa = 4.7FLAPTGEE241 pKa = 4.32SALSEE246 pKa = 3.58IAVFNIEE253 pKa = 4.34GGNITTFDD261 pKa = 3.56EE262 pKa = 4.51
MM1 pKa = 7.51NRR3 pKa = 11.84FCRR6 pKa = 11.84ILRR9 pKa = 11.84QAFAVLLMTFIIAGGGYY26 pKa = 10.33GEE28 pKa = 4.28AVKK31 pKa = 10.86AGWCFAASLDD41 pKa = 4.45LAGLEE46 pKa = 4.38DD47 pKa = 3.94RR48 pKa = 11.84AVDD51 pKa = 3.8IDD53 pKa = 4.05PSDD56 pKa = 3.8GSVSVTLSEE65 pKa = 4.95SEE67 pKa = 4.22MEE69 pKa = 4.21LLSSVRR75 pKa = 11.84CNLLYY80 pKa = 10.59LDD82 pKa = 4.35EE83 pKa = 4.72NVVIHH88 pKa = 6.76LGSDD92 pKa = 3.31ANIDD96 pKa = 3.88ADD98 pKa = 3.21WDD100 pKa = 3.89TGVFRR105 pKa = 11.84DD106 pKa = 4.38NFDD109 pKa = 3.67GTWPMLDD116 pKa = 3.2GHH118 pKa = 6.32PVYY121 pKa = 10.52IEE123 pKa = 3.88LTEE126 pKa = 4.54EE127 pKa = 4.38GDD129 pKa = 3.99DD130 pKa = 3.76YY131 pKa = 11.75NLYY134 pKa = 10.6SIPIKK139 pKa = 10.96LNGEE143 pKa = 3.91EE144 pKa = 5.07CILQVSYY151 pKa = 11.25NYY153 pKa = 10.57NDD155 pKa = 3.36EE156 pKa = 4.44TYY158 pKa = 10.2HH159 pKa = 7.63ILGAQRR165 pKa = 11.84EE166 pKa = 4.32PEE168 pKa = 4.01EE169 pKa = 5.34SGMSDD174 pKa = 2.86QGLIPLRR181 pKa = 11.84EE182 pKa = 4.04GDD184 pKa = 3.99MVTTIHH190 pKa = 6.27FAKK193 pKa = 10.4SVSDD197 pKa = 3.85NDD199 pKa = 3.9DD200 pKa = 4.01DD201 pKa = 4.1EE202 pKa = 4.83LAAVEE207 pKa = 5.19AEE209 pKa = 4.12TFQIGSHH216 pKa = 6.32PAVEE220 pKa = 4.47DD221 pKa = 3.82EE222 pKa = 4.21EE223 pKa = 6.35LGDD226 pKa = 3.59GTYY229 pKa = 10.9GYY231 pKa = 9.91IFEE234 pKa = 4.7FLAPTGEE241 pKa = 4.32SALSEE246 pKa = 3.58IAVFNIEE253 pKa = 4.34GGNITTFDD261 pKa = 3.56EE262 pKa = 4.51
Molecular weight: 28.77 kDa
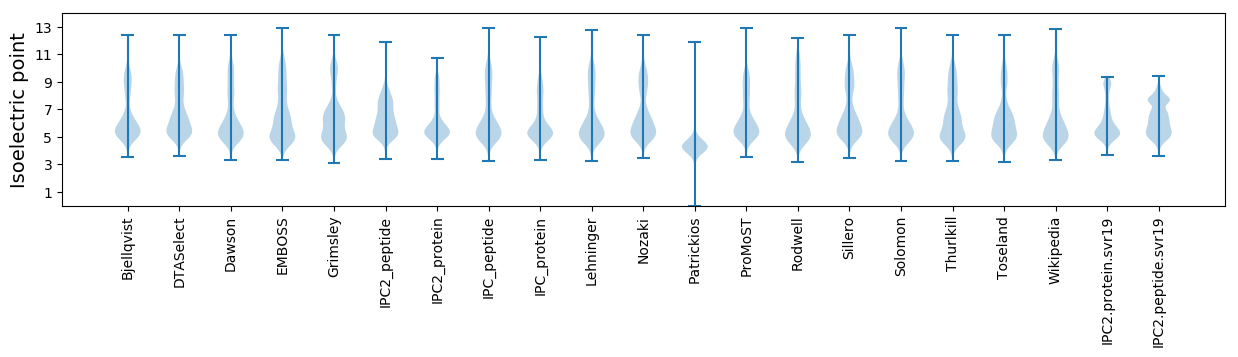
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C9LRM5|C9LRM5_SELS3 RNA polymerase sigma factor sigma-70 family OS=Selenomonas sputigena (strain ATCC 35185 / DSM 20758 / VPI D19B-28) OX=546271 GN=Selsp_1911 PE=4 SV=1
MM1 pKa = 7.22KK2 pKa = 10.11HH3 pKa = 5.81WDD5 pKa = 3.99LPEE8 pKa = 4.43DD9 pKa = 3.84WEE11 pKa = 4.7IEE13 pKa = 4.09AALPEE18 pKa = 4.36YY19 pKa = 11.13VDD21 pKa = 4.1GEE23 pKa = 4.38GDD25 pKa = 3.3ATRR28 pKa = 11.84LFLADD33 pKa = 3.15GTDD36 pKa = 2.97RR37 pKa = 11.84LVRR40 pKa = 11.84ARR42 pKa = 11.84LRR44 pKa = 11.84TVLEE48 pKa = 3.79RR49 pKa = 11.84LARR52 pKa = 11.84RR53 pKa = 11.84HH54 pKa = 5.9CRR56 pKa = 11.84SLPLLRR62 pKa = 11.84AWAKK66 pKa = 10.28EE67 pKa = 3.82RR68 pKa = 11.84TALAQTAPLAIASEE82 pKa = 4.28LVLVPFRR89 pKa = 11.84ARR91 pKa = 11.84RR92 pKa = 11.84PRR94 pKa = 11.84IRR96 pKa = 11.84GDD98 pKa = 2.87AAMGVVNAMHH108 pKa = 6.4AQLVRR113 pKa = 11.84AEE115 pKa = 4.23EE116 pKa = 3.9PAEE119 pKa = 4.1IEE121 pKa = 4.17LSCGRR126 pKa = 11.84RR127 pKa = 11.84IRR129 pKa = 11.84ALWGAATLAAHH140 pKa = 6.75LRR142 pKa = 11.84AAEE145 pKa = 4.32ILRR148 pKa = 11.84QDD150 pKa = 4.06LDD152 pKa = 3.8AKK154 pKa = 10.33TEE156 pKa = 3.98AAVLRR161 pKa = 11.84RR162 pKa = 11.84LFWQKK167 pKa = 9.79AWNRR171 pKa = 11.84AALGRR176 pKa = 11.84KK177 pKa = 8.43
MM1 pKa = 7.22KK2 pKa = 10.11HH3 pKa = 5.81WDD5 pKa = 3.99LPEE8 pKa = 4.43DD9 pKa = 3.84WEE11 pKa = 4.7IEE13 pKa = 4.09AALPEE18 pKa = 4.36YY19 pKa = 11.13VDD21 pKa = 4.1GEE23 pKa = 4.38GDD25 pKa = 3.3ATRR28 pKa = 11.84LFLADD33 pKa = 3.15GTDD36 pKa = 2.97RR37 pKa = 11.84LVRR40 pKa = 11.84ARR42 pKa = 11.84LRR44 pKa = 11.84TVLEE48 pKa = 3.79RR49 pKa = 11.84LARR52 pKa = 11.84RR53 pKa = 11.84HH54 pKa = 5.9CRR56 pKa = 11.84SLPLLRR62 pKa = 11.84AWAKK66 pKa = 10.28EE67 pKa = 3.82RR68 pKa = 11.84TALAQTAPLAIASEE82 pKa = 4.28LVLVPFRR89 pKa = 11.84ARR91 pKa = 11.84RR92 pKa = 11.84PRR94 pKa = 11.84IRR96 pKa = 11.84GDD98 pKa = 2.87AAMGVVNAMHH108 pKa = 6.4AQLVRR113 pKa = 11.84AEE115 pKa = 4.23EE116 pKa = 3.9PAEE119 pKa = 4.1IEE121 pKa = 4.17LSCGRR126 pKa = 11.84RR127 pKa = 11.84IRR129 pKa = 11.84ALWGAATLAAHH140 pKa = 6.75LRR142 pKa = 11.84AAEE145 pKa = 4.32ILRR148 pKa = 11.84QDD150 pKa = 4.06LDD152 pKa = 3.8AKK154 pKa = 10.33TEE156 pKa = 3.98AAVLRR161 pKa = 11.84RR162 pKa = 11.84LFWQKK167 pKa = 9.79AWNRR171 pKa = 11.84AALGRR176 pKa = 11.84KK177 pKa = 8.43
Molecular weight: 20.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
752766 |
32 |
4439 |
335.6 |
37.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.777 ± 0.081 | 1.133 ± 0.023 |
5.453 ± 0.04 | 7.513 ± 0.072 |
3.992 ± 0.045 | 7.69 ± 0.081 |
2.056 ± 0.021 | 5.811 ± 0.057 |
5.604 ± 0.044 | 9.721 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.826 ± 0.029 | 3.128 ± 0.059 |
3.731 ± 0.033 | 2.977 ± 0.031 |
5.906 ± 0.057 | 5.311 ± 0.039 |
5.137 ± 0.081 | 7.14 ± 0.045 |
0.935 ± 0.022 | 3.16 ± 0.043 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
