
Mosquito VEM Anellovirus SDBVL A
Taxonomy: Viruses; Anelloviridae; unclassified Anelloviridae
Average proteome isoelectric point is 9.32
Get precalculated fractions of proteins
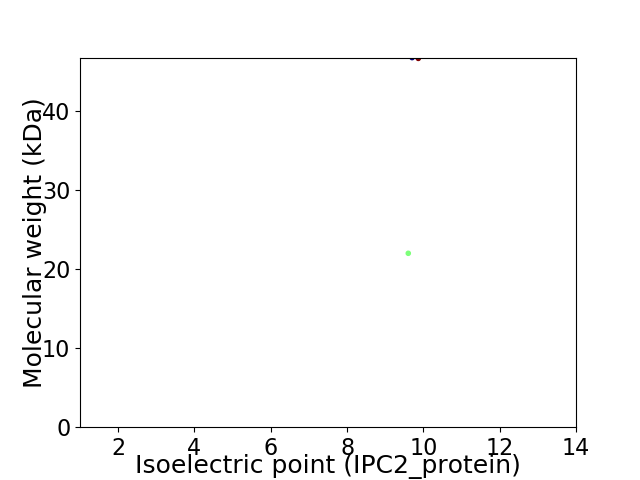
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F6KID0|F6KID0_9VIRU Uncharacterized protein OS=Mosquito VEM Anellovirus SDBVL A OX=1034790 PE=4 SV=1
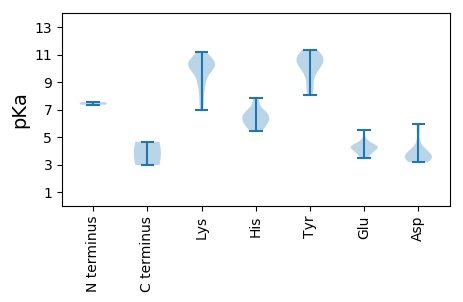
MM1 pKa = 7.54ALYY4 pKa = 9.95FRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84TWRR13 pKa = 11.84PRR15 pKa = 11.84WRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84GWWRR23 pKa = 11.84RR24 pKa = 11.84SYY26 pKa = 10.64RR27 pKa = 11.84SAYY30 pKa = 8.58GRR32 pKa = 11.84RR33 pKa = 11.84FRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84TRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84SRR45 pKa = 11.84VRR47 pKa = 11.84VRR49 pKa = 11.84VLTEE53 pKa = 3.47RR54 pKa = 11.84QPRR57 pKa = 11.84YY58 pKa = 8.33WRR60 pKa = 11.84RR61 pKa = 11.84CTIRR65 pKa = 11.84GWWPVLITGRR75 pKa = 11.84KK76 pKa = 7.55YY77 pKa = 10.69PSTAAQGDD85 pKa = 3.93DD86 pKa = 4.53AGTQMLTFKK95 pKa = 10.27PFQPFGSKK103 pKa = 10.44LSNAWFYY110 pKa = 11.36RR111 pKa = 11.84DD112 pKa = 3.61YY113 pKa = 11.13EE114 pKa = 4.19IVLGGFSAGTFSLQALYY131 pKa = 9.63MEE133 pKa = 4.61HH134 pKa = 7.81LMYY137 pKa = 10.7RR138 pKa = 11.84NRR140 pKa = 11.84WSQSNCGFDD149 pKa = 3.28LASYY153 pKa = 10.31RR154 pKa = 11.84GTKK157 pKa = 10.1LYY159 pKa = 10.27FLPHH163 pKa = 6.78PNYY166 pKa = 10.77DD167 pKa = 4.13YY168 pKa = 11.13IVLIDD173 pKa = 3.4PEE175 pKa = 4.31YY176 pKa = 11.01RR177 pKa = 11.84KK178 pKa = 10.14FEE180 pKa = 3.91QWIKK184 pKa = 10.76QPMHH188 pKa = 6.55PGVLITHH195 pKa = 6.17PQSRR199 pKa = 11.84IIRR202 pKa = 11.84SISHH206 pKa = 6.71AGPRR210 pKa = 11.84RR211 pKa = 11.84KK212 pKa = 8.76MPKK215 pKa = 8.94MFVPPPSTMNSGWQWMGEE233 pKa = 4.03LANEE237 pKa = 4.3GLFAFYY243 pKa = 10.2VQWIDD248 pKa = 5.72LGAPWLGDD256 pKa = 3.3VQNPNQVKK264 pKa = 8.2WWKK267 pKa = 9.23TGDD270 pKa = 3.42ATTKK274 pKa = 10.03PDD276 pKa = 3.3WVTKK280 pKa = 10.07SIEE283 pKa = 4.2MQSKK287 pKa = 7.61TVQEE291 pKa = 4.43GLDD294 pKa = 3.53TYY296 pKa = 10.63YY297 pKa = 11.23SRR299 pKa = 11.84AGTGDD304 pKa = 3.97KK305 pKa = 10.56KK306 pKa = 10.24WVDD309 pKa = 3.81PGSAPYY315 pKa = 10.41QLGFGPFVFKK325 pKa = 10.8GCHH328 pKa = 5.81TGDD331 pKa = 3.48QEE333 pKa = 4.91HH334 pKa = 6.51YY335 pKa = 8.96PQITFFYY342 pKa = 10.08KK343 pKa = 10.48SYY345 pKa = 8.07WQWGGSTTSIKK356 pKa = 9.26TVCDD360 pKa = 3.6PKK362 pKa = 11.21DD363 pKa = 3.68NPNAEE368 pKa = 4.48YY369 pKa = 10.67KK370 pKa = 10.42SAWNRR375 pKa = 11.84AGFKK379 pKa = 10.32PEE381 pKa = 3.54QARR384 pKa = 11.84NSDD387 pKa = 4.05IYY389 pKa = 10.76NQPP392 pKa = 2.97
MM1 pKa = 7.54ALYY4 pKa = 9.95FRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84TWRR13 pKa = 11.84PRR15 pKa = 11.84WRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84GWWRR23 pKa = 11.84RR24 pKa = 11.84SYY26 pKa = 10.64RR27 pKa = 11.84SAYY30 pKa = 8.58GRR32 pKa = 11.84RR33 pKa = 11.84FRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84TRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84SRR45 pKa = 11.84VRR47 pKa = 11.84VRR49 pKa = 11.84VLTEE53 pKa = 3.47RR54 pKa = 11.84QPRR57 pKa = 11.84YY58 pKa = 8.33WRR60 pKa = 11.84RR61 pKa = 11.84CTIRR65 pKa = 11.84GWWPVLITGRR75 pKa = 11.84KK76 pKa = 7.55YY77 pKa = 10.69PSTAAQGDD85 pKa = 3.93DD86 pKa = 4.53AGTQMLTFKK95 pKa = 10.27PFQPFGSKK103 pKa = 10.44LSNAWFYY110 pKa = 11.36RR111 pKa = 11.84DD112 pKa = 3.61YY113 pKa = 11.13EE114 pKa = 4.19IVLGGFSAGTFSLQALYY131 pKa = 9.63MEE133 pKa = 4.61HH134 pKa = 7.81LMYY137 pKa = 10.7RR138 pKa = 11.84NRR140 pKa = 11.84WSQSNCGFDD149 pKa = 3.28LASYY153 pKa = 10.31RR154 pKa = 11.84GTKK157 pKa = 10.1LYY159 pKa = 10.27FLPHH163 pKa = 6.78PNYY166 pKa = 10.77DD167 pKa = 4.13YY168 pKa = 11.13IVLIDD173 pKa = 3.4PEE175 pKa = 4.31YY176 pKa = 11.01RR177 pKa = 11.84KK178 pKa = 10.14FEE180 pKa = 3.91QWIKK184 pKa = 10.76QPMHH188 pKa = 6.55PGVLITHH195 pKa = 6.17PQSRR199 pKa = 11.84IIRR202 pKa = 11.84SISHH206 pKa = 6.71AGPRR210 pKa = 11.84RR211 pKa = 11.84KK212 pKa = 8.76MPKK215 pKa = 8.94MFVPPPSTMNSGWQWMGEE233 pKa = 4.03LANEE237 pKa = 4.3GLFAFYY243 pKa = 10.2VQWIDD248 pKa = 5.72LGAPWLGDD256 pKa = 3.3VQNPNQVKK264 pKa = 8.2WWKK267 pKa = 9.23TGDD270 pKa = 3.42ATTKK274 pKa = 10.03PDD276 pKa = 3.3WVTKK280 pKa = 10.07SIEE283 pKa = 4.2MQSKK287 pKa = 7.61TVQEE291 pKa = 4.43GLDD294 pKa = 3.53TYY296 pKa = 10.63YY297 pKa = 11.23SRR299 pKa = 11.84AGTGDD304 pKa = 3.97KK305 pKa = 10.56KK306 pKa = 10.24WVDD309 pKa = 3.81PGSAPYY315 pKa = 10.41QLGFGPFVFKK325 pKa = 10.8GCHH328 pKa = 5.81TGDD331 pKa = 3.48QEE333 pKa = 4.91HH334 pKa = 6.51YY335 pKa = 8.96PQITFFYY342 pKa = 10.08KK343 pKa = 10.48SYY345 pKa = 8.07WQWGGSTTSIKK356 pKa = 9.26TVCDD360 pKa = 3.6PKK362 pKa = 11.21DD363 pKa = 3.68NPNAEE368 pKa = 4.48YY369 pKa = 10.67KK370 pKa = 10.42SAWNRR375 pKa = 11.84AGFKK379 pKa = 10.32PEE381 pKa = 3.54QARR384 pKa = 11.84NSDD387 pKa = 4.05IYY389 pKa = 10.76NQPP392 pKa = 2.97
Molecular weight: 46.61 kDa
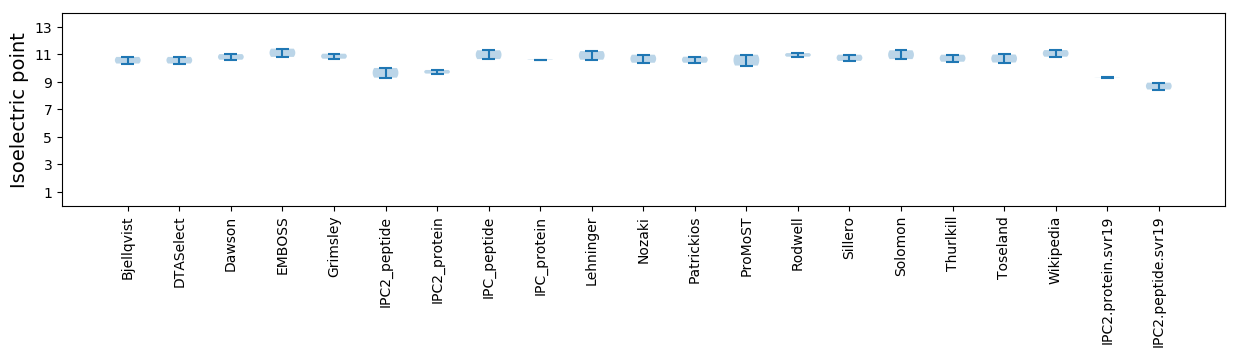
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F6KID0|F6KID0_9VIRU Uncharacterized protein OS=Mosquito VEM Anellovirus SDBVL A OX=1034790 PE=4 SV=1
MM1 pKa = 7.35QNTNRR6 pKa = 11.84PGIGLGLNLSRR17 pKa = 11.84PEE19 pKa = 3.61IVTFIINHH27 pKa = 5.47RR28 pKa = 11.84PKK30 pKa = 9.94VTFDD34 pKa = 3.76LSPPEE39 pKa = 4.34LEE41 pKa = 4.22PVQTSTQTRR50 pKa = 11.84SGRR53 pKa = 11.84PLKK56 pKa = 10.34RR57 pKa = 11.84RR58 pKa = 11.84SIVRR62 pKa = 11.84MQKK65 pKa = 6.99TTTSGGSSIQTPFKK79 pKa = 10.73EE80 pKa = 4.16LLNKK84 pKa = 10.35LSEE87 pKa = 4.41SQCRR91 pKa = 11.84QLASALFPKK100 pKa = 10.24SEE102 pKa = 3.97SRR104 pKa = 11.84ATMRR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84RR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84RR114 pKa = 11.84SMLPRR119 pKa = 11.84RR120 pKa = 11.84KK121 pKa = 8.84RR122 pKa = 11.84TRR124 pKa = 11.84EE125 pKa = 3.86HH126 pKa = 5.99EE127 pKa = 4.18TDD129 pKa = 3.16SDD131 pKa = 3.57EE132 pKa = 5.52GSFGDD137 pKa = 3.45SGSYY141 pKa = 9.91SSRR144 pKa = 11.84GGGKK148 pKa = 9.87RR149 pKa = 11.84SGSEE153 pKa = 3.8DD154 pKa = 3.35TSGSEE159 pKa = 4.33SSSGLDD165 pKa = 5.14DD166 pKa = 5.91PDD168 pKa = 3.98TEE170 pKa = 4.35QNYY173 pKa = 9.36PPVRR177 pKa = 11.84TRR179 pKa = 11.84LAPIKK184 pKa = 10.09FSKK187 pKa = 10.31DD188 pKa = 3.33CRR190 pKa = 11.84PLGPWMM196 pKa = 4.66
MM1 pKa = 7.35QNTNRR6 pKa = 11.84PGIGLGLNLSRR17 pKa = 11.84PEE19 pKa = 3.61IVTFIINHH27 pKa = 5.47RR28 pKa = 11.84PKK30 pKa = 9.94VTFDD34 pKa = 3.76LSPPEE39 pKa = 4.34LEE41 pKa = 4.22PVQTSTQTRR50 pKa = 11.84SGRR53 pKa = 11.84PLKK56 pKa = 10.34RR57 pKa = 11.84RR58 pKa = 11.84SIVRR62 pKa = 11.84MQKK65 pKa = 6.99TTTSGGSSIQTPFKK79 pKa = 10.73EE80 pKa = 4.16LLNKK84 pKa = 10.35LSEE87 pKa = 4.41SQCRR91 pKa = 11.84QLASALFPKK100 pKa = 10.24SEE102 pKa = 3.97SRR104 pKa = 11.84ATMRR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84RR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84RR114 pKa = 11.84SMLPRR119 pKa = 11.84RR120 pKa = 11.84KK121 pKa = 8.84RR122 pKa = 11.84TRR124 pKa = 11.84EE125 pKa = 3.86HH126 pKa = 5.99EE127 pKa = 4.18TDD129 pKa = 3.16SDD131 pKa = 3.57EE132 pKa = 5.52GSFGDD137 pKa = 3.45SGSYY141 pKa = 9.91SSRR144 pKa = 11.84GGGKK148 pKa = 9.87RR149 pKa = 11.84SGSEE153 pKa = 3.8DD154 pKa = 3.35TSGSEE159 pKa = 4.33SSSGLDD165 pKa = 5.14DD166 pKa = 5.91PDD168 pKa = 3.98TEE170 pKa = 4.35QNYY173 pKa = 9.36PPVRR177 pKa = 11.84TRR179 pKa = 11.84LAPIKK184 pKa = 10.09FSKK187 pKa = 10.31DD188 pKa = 3.33CRR190 pKa = 11.84PLGPWMM196 pKa = 4.66
Molecular weight: 21.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
588 |
196 |
392 |
294.0 |
34.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.082 ± 1.083 | 1.02 ± 0.0 |
4.422 ± 0.09 | 4.082 ± 1.083 |
4.422 ± 0.722 | 7.993 ± 0.09 |
1.531 ± 0.271 | 3.741 ± 0.09 |
5.272 ± 0.09 | 5.952 ± 0.902 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.551 ± 0.0 | 3.231 ± 0.09 |
7.483 ± 0.361 | 4.932 ± 0.451 |
11.905 ± 0.722 | 9.014 ± 2.797 |
6.803 ± 0.722 | 3.571 ± 0.541 |
3.741 ± 1.714 | 4.252 ± 1.714 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
