
Amylibacter cionae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Amylibacter
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
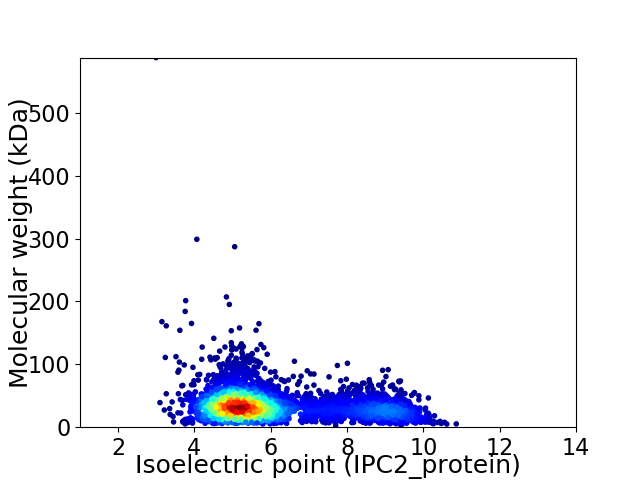
Virtual 2D-PAGE plot for 3948 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N5ILU8|A0A2N5ILU8_9RHOB Guanine deaminase OS=Amylibacter cionae OX=2035344 GN=C0U40_02015 PE=4 SV=1
MM1 pKa = 6.95VRR3 pKa = 11.84STTFEE8 pKa = 4.44FICMPMITHH17 pKa = 6.53TNDD20 pKa = 3.23RR21 pKa = 11.84ARR23 pKa = 11.84KK24 pKa = 8.55AAITPRR30 pKa = 11.84AACMRR35 pKa = 11.84SSAHH39 pKa = 5.68CFARR43 pKa = 11.84LFVRR47 pKa = 11.84YY48 pKa = 9.24SVLLLLAVLFALVKK62 pKa = 10.76SGGVWAQQLILNPTGAPTVVGNGIGKK88 pKa = 9.78RR89 pKa = 11.84GVWRR93 pKa = 11.84NVGTVNGASVDD104 pKa = 3.21IVGIMTLATRR114 pKa = 11.84DD115 pKa = 3.52HH116 pKa = 7.08RR117 pKa = 11.84FGTGNGQIMITSVGQDD133 pKa = 2.89PHH135 pKa = 7.15FADD138 pKa = 3.93FHH140 pKa = 8.08IYY142 pKa = 9.64EE143 pKa = 4.77AGTYY147 pKa = 10.43NIFSNSGGVPVVADD161 pKa = 3.46VDD163 pKa = 3.77IQINDD168 pKa = 2.88IDD170 pKa = 4.54GPVNEE175 pKa = 4.08QVYY178 pKa = 11.16VNLCDD183 pKa = 4.03GSVEE187 pKa = 4.1YY188 pKa = 11.21VRR190 pKa = 11.84VDD192 pKa = 3.76RR193 pKa = 11.84NATTYY198 pKa = 10.15RR199 pKa = 11.84GYY201 pKa = 10.32IEE203 pKa = 5.93GPDD206 pKa = 3.51ANLGTEE212 pKa = 4.15VFYY215 pKa = 11.03LAGDD219 pKa = 3.45QNYY222 pKa = 10.42ANQPVSGLEE231 pKa = 3.3ISYY234 pKa = 9.06PQTSTFSFGRR244 pKa = 11.84TANNGFLVLLANPTYY259 pKa = 10.47DD260 pKa = 3.79VSDD263 pKa = 3.83TYY265 pKa = 10.89EE266 pKa = 4.18LKK268 pKa = 11.09CGDD271 pKa = 3.82FKK273 pKa = 11.78APVLQDD279 pKa = 4.14DD280 pKa = 4.18IKK282 pKa = 10.62EE283 pKa = 4.19QVLGEE288 pKa = 4.11PVTVNILFNDD298 pKa = 3.86SVATEE303 pKa = 4.22NDD305 pKa = 2.95NAPANNSQQPSEE317 pKa = 4.23YY318 pKa = 10.95AMQAIDD324 pKa = 5.75LIPPTGALNIVTDD337 pKa = 4.12SAGHH341 pKa = 5.77RR342 pKa = 11.84VGFDD346 pKa = 3.35VIGEE350 pKa = 4.49GTWSYY355 pKa = 12.03DD356 pKa = 3.37DD357 pKa = 3.55VTGEE361 pKa = 3.95LTFTPFVAFFAAPTPIDD378 pKa = 3.53YY379 pKa = 10.5RR380 pKa = 11.84FQSPIILPSEE390 pKa = 4.04PQAYY394 pKa = 8.9SAPARR399 pKa = 11.84VSIDD403 pKa = 3.36VGSVGLLKK411 pKa = 10.54LAQLVDD417 pKa = 3.88TNLNGYY423 pKa = 9.4ADD425 pKa = 4.32PGEE428 pKa = 4.36TIAYY432 pKa = 9.14VFTAEE437 pKa = 4.1NFGNVDD443 pKa = 3.57LTNVQLAEE451 pKa = 4.29TQFSGAGIPPVITFQAATSLSPEE474 pKa = 3.84GTLLVGEE481 pKa = 4.4KK482 pKa = 10.26AVYY485 pKa = 7.91TATYY489 pKa = 9.11TLVPEE494 pKa = 5.2DD495 pKa = 4.76LDD497 pKa = 4.03TTISNQAEE505 pKa = 4.34VTAQTPSGTTVSDD518 pKa = 3.93LSDD521 pKa = 3.67SEE523 pKa = 4.42NPSDD527 pKa = 3.88GDD529 pKa = 3.71GVARR533 pKa = 11.84NGPGEE538 pKa = 4.19GRR540 pKa = 11.84DD541 pKa = 3.91DD542 pKa = 3.61PTTIYY547 pKa = 10.61AGSGPDD553 pKa = 4.01RR554 pKa = 11.84GDD556 pKa = 3.35APITYY561 pKa = 10.05GDD563 pKa = 4.06PQHH566 pKa = 7.14ADD568 pKa = 3.04TSAYY572 pKa = 9.16WIGALNGDD580 pKa = 3.97GDD582 pKa = 4.12GSAQHH587 pKa = 6.48SADD590 pKa = 3.42GTGDD594 pKa = 3.87DD595 pKa = 5.81LDD597 pKa = 4.75GKK599 pKa = 10.5DD600 pKa = 5.28DD601 pKa = 3.83EE602 pKa = 5.08SEE604 pKa = 4.26EE605 pKa = 4.39NFPQLYY611 pKa = 10.66GDD613 pKa = 4.16LTRR616 pKa = 11.84TVTIPVNEE624 pKa = 4.04PVPGSGYY631 pKa = 10.14LQAFIDD637 pKa = 3.98FGGDD641 pKa = 3.21GTFLSLGDD649 pKa = 3.82QVATDD654 pKa = 3.73IQDD657 pKa = 3.95GGPQDD662 pKa = 4.92LDD664 pKa = 3.34GTANGEE670 pKa = 3.64ISFAISVPATAVLTPTFARR689 pKa = 11.84LRR691 pKa = 11.84WSSVAGLTAISPATDD706 pKa = 3.7GEE708 pKa = 4.46VEE710 pKa = 4.76DD711 pKa = 4.26YY712 pKa = 11.18GITIKK717 pKa = 10.01TPPEE721 pKa = 4.08ADD723 pKa = 3.59RR724 pKa = 11.84GDD726 pKa = 3.83APASYY731 pKa = 10.79GDD733 pKa = 3.62PQHH736 pKa = 7.29IVEE739 pKa = 5.2GPAAPEE745 pKa = 4.09IYY747 pKa = 10.21LGTIAPDD754 pKa = 3.09VDD756 pKa = 4.85VIAQNTATASGDD768 pKa = 3.82DD769 pKa = 3.97LDD771 pKa = 6.05GNDD774 pKa = 4.88DD775 pKa = 3.57EE776 pKa = 6.91DD777 pKa = 4.91GVVLPTLYY785 pKa = 10.86LGGLAEE791 pKa = 4.29ITVTVNDD798 pKa = 4.0VGSPPQKK805 pKa = 10.04SAYY808 pKa = 9.12LQAFIDD814 pKa = 3.88FDD816 pKa = 4.63GDD818 pKa = 3.84GTFLQAGEE826 pKa = 4.18QVATNLQDD834 pKa = 4.65GDD836 pKa = 4.33PLDD839 pKa = 4.53KK840 pKa = 11.24DD841 pKa = 3.94GASDD845 pKa = 3.54GSIRR849 pKa = 11.84FEE851 pKa = 4.14VAVPAGATTLPTFARR866 pKa = 11.84FRR868 pKa = 11.84WSTDD872 pKa = 2.48ATGQAIAFNGEE883 pKa = 3.93VEE885 pKa = 5.09DD886 pKa = 4.21YY887 pKa = 10.38PLTISGDD894 pKa = 3.96PPPFLCDD901 pKa = 3.45GSIYY905 pKa = 10.24RR906 pKa = 11.84YY907 pKa = 10.6DD908 pKa = 3.71DD909 pKa = 3.48DD910 pKa = 4.36RR911 pKa = 11.84VVRR914 pKa = 11.84RR915 pKa = 11.84LTFTASGSSYY925 pKa = 11.14AIGFQDD931 pKa = 4.02IGTAPRR937 pKa = 11.84NLNANWGYY945 pKa = 10.52NALDD949 pKa = 3.84GYY951 pKa = 9.97MYY953 pKa = 10.85AVRR956 pKa = 11.84PGRR959 pKa = 11.84SDD961 pKa = 4.29LYY963 pKa = 11.11RR964 pKa = 11.84LDD966 pKa = 3.68GAGNFVDD973 pKa = 4.74LTNLPGGTADD983 pKa = 3.76GSGAGDD989 pKa = 3.57IMPNGTMVYY998 pKa = 9.68VANGNTWQLISLVDD1012 pKa = 3.6ASNPVNLGVLNLSQSVDD1029 pKa = 3.4TRR1031 pKa = 11.84DD1032 pKa = 3.12IAYY1035 pKa = 10.35NPVDD1039 pKa = 3.33GLFYY1043 pKa = 10.83GINQSSGRR1051 pKa = 11.84AFKK1054 pKa = 10.68VDD1056 pKa = 3.69PNGGVAGLATVTEE1069 pKa = 5.06FGPAIYY1075 pKa = 10.12GGIFGSVWFDD1085 pKa = 3.02QDD1087 pKa = 2.69GRR1089 pKa = 11.84FYY1091 pKa = 11.22VYY1093 pKa = 11.24SNTTNNLYY1101 pKa = 8.83LTNTTTGEE1109 pKa = 3.9AQLIANSTVDD1119 pKa = 3.39EE1120 pKa = 5.09AGDD1123 pKa = 3.86SDD1125 pKa = 4.32GMSCRR1130 pKa = 11.84GPAPIPFGAISGNVYY1145 pKa = 10.99DD1146 pKa = 5.01DD1147 pKa = 4.3TNASDD1152 pKa = 3.81VKK1154 pKa = 11.49DD1155 pKa = 3.59NGEE1158 pKa = 4.23TNLGAGIAISVYY1170 pKa = 10.74ADD1172 pKa = 2.98SGTPADD1178 pKa = 3.9TSDD1181 pKa = 4.29DD1182 pKa = 3.91LFLITTDD1189 pKa = 3.44TEE1191 pKa = 4.34ADD1193 pKa = 3.15GTYY1196 pKa = 10.89AVADD1200 pKa = 3.83LLINTNYY1207 pKa = 10.0RR1208 pKa = 11.84VEE1210 pKa = 4.54LDD1212 pKa = 3.74EE1213 pKa = 5.96ADD1215 pKa = 4.01PDD1217 pKa = 3.87LGGGKK1222 pKa = 8.45TIGTSNPRR1230 pKa = 11.84LGVEE1234 pKa = 4.14VTAGEE1239 pKa = 4.43TTTDD1243 pKa = 2.84QDD1245 pKa = 3.92FGFDD1249 pKa = 3.3AAASDD1254 pKa = 4.98LEE1256 pKa = 4.32LTKK1259 pKa = 10.95YY1260 pKa = 10.49AALTGTTTPVTNVSEE1275 pKa = 4.32GDD1277 pKa = 3.7VIDD1280 pKa = 4.41WIITITNVSGGSPSGVKK1297 pKa = 10.5VIDD1300 pKa = 5.25LIPSGFAYY1308 pKa = 10.72VSDD1311 pKa = 4.3SAPSTGDD1318 pKa = 2.92TYY1320 pKa = 11.76DD1321 pKa = 4.56PDD1323 pKa = 3.71TGLWFVDD1330 pKa = 4.89EE1331 pKa = 4.45ILAGASEE1338 pKa = 4.4TLVITTTVLDD1348 pKa = 3.88SGEE1351 pKa = 4.17LTNTAEE1357 pKa = 4.8IIYY1360 pKa = 10.41SSLPDD1365 pKa = 4.36PDD1367 pKa = 4.99SDD1369 pKa = 4.19PSTGPLTDD1377 pKa = 4.97DD1378 pKa = 5.15LFDD1381 pKa = 4.82QIADD1385 pKa = 4.04DD1386 pKa = 5.38DD1387 pKa = 4.35EE1388 pKa = 6.53ASYY1391 pKa = 9.74TVNLITNDD1399 pKa = 3.24RR1400 pKa = 11.84VFSGRR1405 pKa = 11.84IFSDD1409 pKa = 2.97NGEE1412 pKa = 4.46SGGTAHH1418 pKa = 7.65DD1419 pKa = 3.89AEE1421 pKa = 4.76INGGEE1426 pKa = 4.21VGLSSALLEE1435 pKa = 4.31IVDD1438 pKa = 3.8NTGTVIATPDD1448 pKa = 3.47VASGGTWTYY1457 pKa = 11.42ALPSSYY1463 pKa = 10.15TGEE1466 pKa = 3.97LTIRR1470 pKa = 11.84AIPANGYY1477 pKa = 9.27RR1478 pKa = 11.84AISEE1482 pKa = 4.28ATNALPGLSNPDD1494 pKa = 3.2PHH1496 pKa = 7.43DD1497 pKa = 3.82GSFTFTPDD1505 pKa = 3.17PSGNLVNLDD1514 pKa = 3.27IGLLEE1519 pKa = 4.51LPILTEE1525 pKa = 4.24DD1526 pKa = 3.3RR1527 pKa = 11.84VSTVGQGQIVALPHH1541 pKa = 7.06LYY1543 pKa = 9.2TATSDD1548 pKa = 3.62GSVTFSLADD1557 pKa = 3.43STSAPTGAFSAALYY1571 pKa = 10.46HH1572 pKa = 7.17DD1573 pKa = 5.26ASCTGAPGVPVTGPLSVSAGEE1594 pKa = 4.37TICIQSRR1601 pKa = 11.84ISAGAGAGTGSRR1613 pKa = 11.84FTYY1616 pKa = 10.17KK1617 pKa = 10.33IIAKK1621 pKa = 6.5TTFTGTTAVSSASNTDD1637 pKa = 3.19QVTVTAEE1644 pKa = 3.78SGQVHH1649 pKa = 5.66LTKK1652 pKa = 10.28TVEE1655 pKa = 3.89NEE1657 pKa = 4.02TAGTAEE1663 pKa = 4.29ATSNMGGANDD1673 pKa = 3.39VLLYY1677 pKa = 10.43RR1678 pKa = 11.84IYY1680 pKa = 11.07LEE1682 pKa = 4.45NTDD1685 pKa = 3.83TTPVTNVKK1693 pKa = 10.27VYY1695 pKa = 11.07DD1696 pKa = 3.58RR1697 pKa = 11.84TPPYY1701 pKa = 9.25TALSEE1706 pKa = 4.8PIADD1710 pKa = 3.97PQILSPNLSCNLAKK1724 pKa = 10.36PVTNTANYY1732 pKa = 9.87SGLVEE1737 pKa = 4.38WNCSGNFLPGEE1748 pKa = 4.08VGALQFRR1755 pKa = 11.84VAITPP1760 pKa = 3.69
MM1 pKa = 6.95VRR3 pKa = 11.84STTFEE8 pKa = 4.44FICMPMITHH17 pKa = 6.53TNDD20 pKa = 3.23RR21 pKa = 11.84ARR23 pKa = 11.84KK24 pKa = 8.55AAITPRR30 pKa = 11.84AACMRR35 pKa = 11.84SSAHH39 pKa = 5.68CFARR43 pKa = 11.84LFVRR47 pKa = 11.84YY48 pKa = 9.24SVLLLLAVLFALVKK62 pKa = 10.76SGGVWAQQLILNPTGAPTVVGNGIGKK88 pKa = 9.78RR89 pKa = 11.84GVWRR93 pKa = 11.84NVGTVNGASVDD104 pKa = 3.21IVGIMTLATRR114 pKa = 11.84DD115 pKa = 3.52HH116 pKa = 7.08RR117 pKa = 11.84FGTGNGQIMITSVGQDD133 pKa = 2.89PHH135 pKa = 7.15FADD138 pKa = 3.93FHH140 pKa = 8.08IYY142 pKa = 9.64EE143 pKa = 4.77AGTYY147 pKa = 10.43NIFSNSGGVPVVADD161 pKa = 3.46VDD163 pKa = 3.77IQINDD168 pKa = 2.88IDD170 pKa = 4.54GPVNEE175 pKa = 4.08QVYY178 pKa = 11.16VNLCDD183 pKa = 4.03GSVEE187 pKa = 4.1YY188 pKa = 11.21VRR190 pKa = 11.84VDD192 pKa = 3.76RR193 pKa = 11.84NATTYY198 pKa = 10.15RR199 pKa = 11.84GYY201 pKa = 10.32IEE203 pKa = 5.93GPDD206 pKa = 3.51ANLGTEE212 pKa = 4.15VFYY215 pKa = 11.03LAGDD219 pKa = 3.45QNYY222 pKa = 10.42ANQPVSGLEE231 pKa = 3.3ISYY234 pKa = 9.06PQTSTFSFGRR244 pKa = 11.84TANNGFLVLLANPTYY259 pKa = 10.47DD260 pKa = 3.79VSDD263 pKa = 3.83TYY265 pKa = 10.89EE266 pKa = 4.18LKK268 pKa = 11.09CGDD271 pKa = 3.82FKK273 pKa = 11.78APVLQDD279 pKa = 4.14DD280 pKa = 4.18IKK282 pKa = 10.62EE283 pKa = 4.19QVLGEE288 pKa = 4.11PVTVNILFNDD298 pKa = 3.86SVATEE303 pKa = 4.22NDD305 pKa = 2.95NAPANNSQQPSEE317 pKa = 4.23YY318 pKa = 10.95AMQAIDD324 pKa = 5.75LIPPTGALNIVTDD337 pKa = 4.12SAGHH341 pKa = 5.77RR342 pKa = 11.84VGFDD346 pKa = 3.35VIGEE350 pKa = 4.49GTWSYY355 pKa = 12.03DD356 pKa = 3.37DD357 pKa = 3.55VTGEE361 pKa = 3.95LTFTPFVAFFAAPTPIDD378 pKa = 3.53YY379 pKa = 10.5RR380 pKa = 11.84FQSPIILPSEE390 pKa = 4.04PQAYY394 pKa = 8.9SAPARR399 pKa = 11.84VSIDD403 pKa = 3.36VGSVGLLKK411 pKa = 10.54LAQLVDD417 pKa = 3.88TNLNGYY423 pKa = 9.4ADD425 pKa = 4.32PGEE428 pKa = 4.36TIAYY432 pKa = 9.14VFTAEE437 pKa = 4.1NFGNVDD443 pKa = 3.57LTNVQLAEE451 pKa = 4.29TQFSGAGIPPVITFQAATSLSPEE474 pKa = 3.84GTLLVGEE481 pKa = 4.4KK482 pKa = 10.26AVYY485 pKa = 7.91TATYY489 pKa = 9.11TLVPEE494 pKa = 5.2DD495 pKa = 4.76LDD497 pKa = 4.03TTISNQAEE505 pKa = 4.34VTAQTPSGTTVSDD518 pKa = 3.93LSDD521 pKa = 3.67SEE523 pKa = 4.42NPSDD527 pKa = 3.88GDD529 pKa = 3.71GVARR533 pKa = 11.84NGPGEE538 pKa = 4.19GRR540 pKa = 11.84DD541 pKa = 3.91DD542 pKa = 3.61PTTIYY547 pKa = 10.61AGSGPDD553 pKa = 4.01RR554 pKa = 11.84GDD556 pKa = 3.35APITYY561 pKa = 10.05GDD563 pKa = 4.06PQHH566 pKa = 7.14ADD568 pKa = 3.04TSAYY572 pKa = 9.16WIGALNGDD580 pKa = 3.97GDD582 pKa = 4.12GSAQHH587 pKa = 6.48SADD590 pKa = 3.42GTGDD594 pKa = 3.87DD595 pKa = 5.81LDD597 pKa = 4.75GKK599 pKa = 10.5DD600 pKa = 5.28DD601 pKa = 3.83EE602 pKa = 5.08SEE604 pKa = 4.26EE605 pKa = 4.39NFPQLYY611 pKa = 10.66GDD613 pKa = 4.16LTRR616 pKa = 11.84TVTIPVNEE624 pKa = 4.04PVPGSGYY631 pKa = 10.14LQAFIDD637 pKa = 3.98FGGDD641 pKa = 3.21GTFLSLGDD649 pKa = 3.82QVATDD654 pKa = 3.73IQDD657 pKa = 3.95GGPQDD662 pKa = 4.92LDD664 pKa = 3.34GTANGEE670 pKa = 3.64ISFAISVPATAVLTPTFARR689 pKa = 11.84LRR691 pKa = 11.84WSSVAGLTAISPATDD706 pKa = 3.7GEE708 pKa = 4.46VEE710 pKa = 4.76DD711 pKa = 4.26YY712 pKa = 11.18GITIKK717 pKa = 10.01TPPEE721 pKa = 4.08ADD723 pKa = 3.59RR724 pKa = 11.84GDD726 pKa = 3.83APASYY731 pKa = 10.79GDD733 pKa = 3.62PQHH736 pKa = 7.29IVEE739 pKa = 5.2GPAAPEE745 pKa = 4.09IYY747 pKa = 10.21LGTIAPDD754 pKa = 3.09VDD756 pKa = 4.85VIAQNTATASGDD768 pKa = 3.82DD769 pKa = 3.97LDD771 pKa = 6.05GNDD774 pKa = 4.88DD775 pKa = 3.57EE776 pKa = 6.91DD777 pKa = 4.91GVVLPTLYY785 pKa = 10.86LGGLAEE791 pKa = 4.29ITVTVNDD798 pKa = 4.0VGSPPQKK805 pKa = 10.04SAYY808 pKa = 9.12LQAFIDD814 pKa = 3.88FDD816 pKa = 4.63GDD818 pKa = 3.84GTFLQAGEE826 pKa = 4.18QVATNLQDD834 pKa = 4.65GDD836 pKa = 4.33PLDD839 pKa = 4.53KK840 pKa = 11.24DD841 pKa = 3.94GASDD845 pKa = 3.54GSIRR849 pKa = 11.84FEE851 pKa = 4.14VAVPAGATTLPTFARR866 pKa = 11.84FRR868 pKa = 11.84WSTDD872 pKa = 2.48ATGQAIAFNGEE883 pKa = 3.93VEE885 pKa = 5.09DD886 pKa = 4.21YY887 pKa = 10.38PLTISGDD894 pKa = 3.96PPPFLCDD901 pKa = 3.45GSIYY905 pKa = 10.24RR906 pKa = 11.84YY907 pKa = 10.6DD908 pKa = 3.71DD909 pKa = 3.48DD910 pKa = 4.36RR911 pKa = 11.84VVRR914 pKa = 11.84RR915 pKa = 11.84LTFTASGSSYY925 pKa = 11.14AIGFQDD931 pKa = 4.02IGTAPRR937 pKa = 11.84NLNANWGYY945 pKa = 10.52NALDD949 pKa = 3.84GYY951 pKa = 9.97MYY953 pKa = 10.85AVRR956 pKa = 11.84PGRR959 pKa = 11.84SDD961 pKa = 4.29LYY963 pKa = 11.11RR964 pKa = 11.84LDD966 pKa = 3.68GAGNFVDD973 pKa = 4.74LTNLPGGTADD983 pKa = 3.76GSGAGDD989 pKa = 3.57IMPNGTMVYY998 pKa = 9.68VANGNTWQLISLVDD1012 pKa = 3.6ASNPVNLGVLNLSQSVDD1029 pKa = 3.4TRR1031 pKa = 11.84DD1032 pKa = 3.12IAYY1035 pKa = 10.35NPVDD1039 pKa = 3.33GLFYY1043 pKa = 10.83GINQSSGRR1051 pKa = 11.84AFKK1054 pKa = 10.68VDD1056 pKa = 3.69PNGGVAGLATVTEE1069 pKa = 5.06FGPAIYY1075 pKa = 10.12GGIFGSVWFDD1085 pKa = 3.02QDD1087 pKa = 2.69GRR1089 pKa = 11.84FYY1091 pKa = 11.22VYY1093 pKa = 11.24SNTTNNLYY1101 pKa = 8.83LTNTTTGEE1109 pKa = 3.9AQLIANSTVDD1119 pKa = 3.39EE1120 pKa = 5.09AGDD1123 pKa = 3.86SDD1125 pKa = 4.32GMSCRR1130 pKa = 11.84GPAPIPFGAISGNVYY1145 pKa = 10.99DD1146 pKa = 5.01DD1147 pKa = 4.3TNASDD1152 pKa = 3.81VKK1154 pKa = 11.49DD1155 pKa = 3.59NGEE1158 pKa = 4.23TNLGAGIAISVYY1170 pKa = 10.74ADD1172 pKa = 2.98SGTPADD1178 pKa = 3.9TSDD1181 pKa = 4.29DD1182 pKa = 3.91LFLITTDD1189 pKa = 3.44TEE1191 pKa = 4.34ADD1193 pKa = 3.15GTYY1196 pKa = 10.89AVADD1200 pKa = 3.83LLINTNYY1207 pKa = 10.0RR1208 pKa = 11.84VEE1210 pKa = 4.54LDD1212 pKa = 3.74EE1213 pKa = 5.96ADD1215 pKa = 4.01PDD1217 pKa = 3.87LGGGKK1222 pKa = 8.45TIGTSNPRR1230 pKa = 11.84LGVEE1234 pKa = 4.14VTAGEE1239 pKa = 4.43TTTDD1243 pKa = 2.84QDD1245 pKa = 3.92FGFDD1249 pKa = 3.3AAASDD1254 pKa = 4.98LEE1256 pKa = 4.32LTKK1259 pKa = 10.95YY1260 pKa = 10.49AALTGTTTPVTNVSEE1275 pKa = 4.32GDD1277 pKa = 3.7VIDD1280 pKa = 4.41WIITITNVSGGSPSGVKK1297 pKa = 10.5VIDD1300 pKa = 5.25LIPSGFAYY1308 pKa = 10.72VSDD1311 pKa = 4.3SAPSTGDD1318 pKa = 2.92TYY1320 pKa = 11.76DD1321 pKa = 4.56PDD1323 pKa = 3.71TGLWFVDD1330 pKa = 4.89EE1331 pKa = 4.45ILAGASEE1338 pKa = 4.4TLVITTTVLDD1348 pKa = 3.88SGEE1351 pKa = 4.17LTNTAEE1357 pKa = 4.8IIYY1360 pKa = 10.41SSLPDD1365 pKa = 4.36PDD1367 pKa = 4.99SDD1369 pKa = 4.19PSTGPLTDD1377 pKa = 4.97DD1378 pKa = 5.15LFDD1381 pKa = 4.82QIADD1385 pKa = 4.04DD1386 pKa = 5.38DD1387 pKa = 4.35EE1388 pKa = 6.53ASYY1391 pKa = 9.74TVNLITNDD1399 pKa = 3.24RR1400 pKa = 11.84VFSGRR1405 pKa = 11.84IFSDD1409 pKa = 2.97NGEE1412 pKa = 4.46SGGTAHH1418 pKa = 7.65DD1419 pKa = 3.89AEE1421 pKa = 4.76INGGEE1426 pKa = 4.21VGLSSALLEE1435 pKa = 4.31IVDD1438 pKa = 3.8NTGTVIATPDD1448 pKa = 3.47VASGGTWTYY1457 pKa = 11.42ALPSSYY1463 pKa = 10.15TGEE1466 pKa = 3.97LTIRR1470 pKa = 11.84AIPANGYY1477 pKa = 9.27RR1478 pKa = 11.84AISEE1482 pKa = 4.28ATNALPGLSNPDD1494 pKa = 3.2PHH1496 pKa = 7.43DD1497 pKa = 3.82GSFTFTPDD1505 pKa = 3.17PSGNLVNLDD1514 pKa = 3.27IGLLEE1519 pKa = 4.51LPILTEE1525 pKa = 4.24DD1526 pKa = 3.3RR1527 pKa = 11.84VSTVGQGQIVALPHH1541 pKa = 7.06LYY1543 pKa = 9.2TATSDD1548 pKa = 3.62GSVTFSLADD1557 pKa = 3.43STSAPTGAFSAALYY1571 pKa = 10.46HH1572 pKa = 7.17DD1573 pKa = 5.26ASCTGAPGVPVTGPLSVSAGEE1594 pKa = 4.37TICIQSRR1601 pKa = 11.84ISAGAGAGTGSRR1613 pKa = 11.84FTYY1616 pKa = 10.17KK1617 pKa = 10.33IIAKK1621 pKa = 6.5TTFTGTTAVSSASNTDD1637 pKa = 3.19QVTVTAEE1644 pKa = 3.78SGQVHH1649 pKa = 5.66LTKK1652 pKa = 10.28TVEE1655 pKa = 3.89NEE1657 pKa = 4.02TAGTAEE1663 pKa = 4.29ATSNMGGANDD1673 pKa = 3.39VLLYY1677 pKa = 10.43RR1678 pKa = 11.84IYY1680 pKa = 11.07LEE1682 pKa = 4.45NTDD1685 pKa = 3.83TTPVTNVKK1693 pKa = 10.27VYY1695 pKa = 11.07DD1696 pKa = 3.58RR1697 pKa = 11.84TPPYY1701 pKa = 9.25TALSEE1706 pKa = 4.8PIADD1710 pKa = 3.97PQILSPNLSCNLAKK1724 pKa = 10.36PVTNTANYY1732 pKa = 9.87SGLVEE1737 pKa = 4.38WNCSGNFLPGEE1748 pKa = 4.08VGALQFRR1755 pKa = 11.84VAITPP1760 pKa = 3.69
Molecular weight: 184.22 kDa
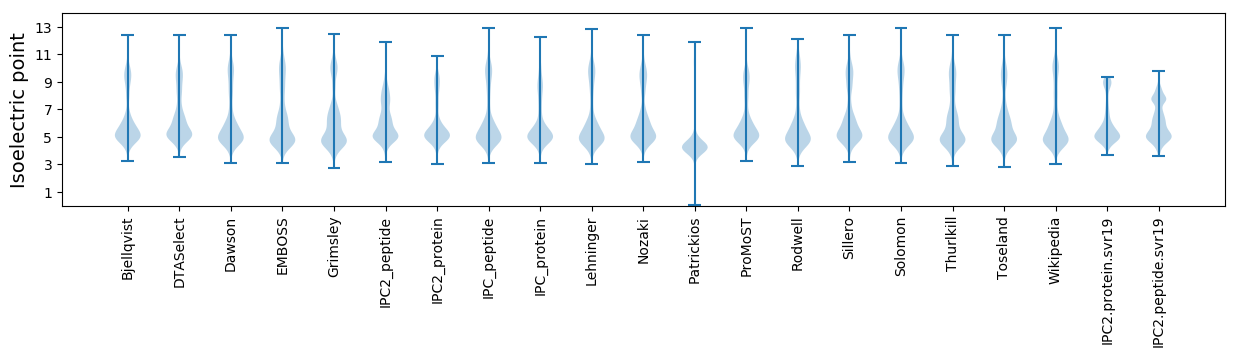
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N5IG79|A0A2N5IG79_9RHOB CoA-binding protein OS=Amylibacter cionae OX=2035344 GN=C0U40_14645 PE=4 SV=1
MM1 pKa = 7.17HH2 pKa = 7.11VRR4 pKa = 11.84RR5 pKa = 11.84TMYY8 pKa = 7.2EE9 pKa = 3.47TSMSGRR15 pKa = 11.84HH16 pKa = 5.95FISQRR21 pKa = 11.84YY22 pKa = 8.07LFRR25 pKa = 11.84SHH27 pKa = 6.91ALSFTLQKK35 pKa = 9.63LTPEE39 pKa = 4.15RR40 pKa = 11.84SKK42 pKa = 11.6SLLARR47 pKa = 11.84AACLPVPIEE56 pKa = 4.47TEE58 pKa = 4.09SQLPTVLHH66 pKa = 5.6VVSAIAANVGIPPFKK81 pKa = 10.41TGFCAATIQHH91 pKa = 6.06QGRR94 pKa = 11.84KK95 pKa = 10.01AEE97 pKa = 4.1VRR99 pKa = 11.84CKK101 pKa = 10.38RR102 pKa = 11.84EE103 pKa = 3.15RR104 pKa = 11.84DD105 pKa = 3.59AILVSKK111 pKa = 10.53RR112 pKa = 11.84AFKK115 pKa = 10.66ACYY118 pKa = 8.39PIPTNNVRR126 pKa = 11.84LL127 pKa = 3.82
MM1 pKa = 7.17HH2 pKa = 7.11VRR4 pKa = 11.84RR5 pKa = 11.84TMYY8 pKa = 7.2EE9 pKa = 3.47TSMSGRR15 pKa = 11.84HH16 pKa = 5.95FISQRR21 pKa = 11.84YY22 pKa = 8.07LFRR25 pKa = 11.84SHH27 pKa = 6.91ALSFTLQKK35 pKa = 9.63LTPEE39 pKa = 4.15RR40 pKa = 11.84SKK42 pKa = 11.6SLLARR47 pKa = 11.84AACLPVPIEE56 pKa = 4.47TEE58 pKa = 4.09SQLPTVLHH66 pKa = 5.6VVSAIAANVGIPPFKK81 pKa = 10.41TGFCAATIQHH91 pKa = 6.06QGRR94 pKa = 11.84KK95 pKa = 10.01AEE97 pKa = 4.1VRR99 pKa = 11.84CKK101 pKa = 10.38RR102 pKa = 11.84EE103 pKa = 3.15RR104 pKa = 11.84DD105 pKa = 3.59AILVSKK111 pKa = 10.53RR112 pKa = 11.84AFKK115 pKa = 10.66ACYY118 pKa = 8.39PIPTNNVRR126 pKa = 11.84LL127 pKa = 3.82
Molecular weight: 14.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1297894 |
21 |
5932 |
328.7 |
35.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.428 ± 0.051 | 0.919 ± 0.012 |
6.058 ± 0.047 | 5.999 ± 0.038 |
3.925 ± 0.029 | 8.587 ± 0.04 |
1.967 ± 0.022 | 5.441 ± 0.028 |
3.796 ± 0.036 | 9.906 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.811 ± 0.026 | 3.004 ± 0.024 |
4.759 ± 0.033 | 3.292 ± 0.021 |
5.883 ± 0.045 | 5.609 ± 0.032 |
5.633 ± 0.036 | 7.342 ± 0.035 |
1.287 ± 0.016 | 2.353 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
