
Enterobacteria phage GA (Bacteriophage GA)
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Leviviricetes; Norzivirales; Fiersviridae; Emesvirus; Emesvirus japonicum
Average proteome isoelectric point is 8.02
Get precalculated fractions of proteins
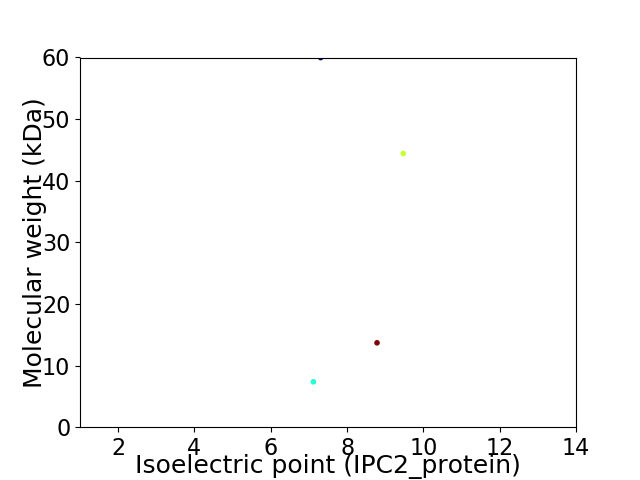
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P07394|MATA_BPGA Maturation protein A OS=Enterobacteria phage GA OX=12018 GN=A PE=2 SV=1
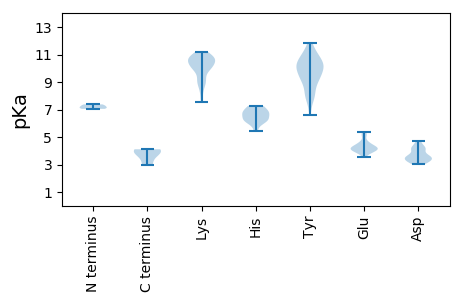
MM1 pKa = 7.12FRR3 pKa = 11.84FRR5 pKa = 11.84EE6 pKa = 4.05IEE8 pKa = 3.77KK9 pKa = 8.89TLCMDD14 pKa = 4.01RR15 pKa = 11.84TRR17 pKa = 11.84DD18 pKa = 3.47CAVRR22 pKa = 11.84FHH24 pKa = 7.27VYY26 pKa = 9.84LQSLDD31 pKa = 4.29LGSSDD36 pKa = 4.25PLSPDD41 pKa = 3.21FDD43 pKa = 3.73GLAYY47 pKa = 10.7LRR49 pKa = 11.84DD50 pKa = 3.68EE51 pKa = 5.36CLTKK55 pKa = 10.59HH56 pKa = 6.39PSLGDD61 pKa = 3.7SNSDD65 pKa = 3.12ARR67 pKa = 11.84RR68 pKa = 11.84KK69 pKa = 9.05EE70 pKa = 3.55LAYY73 pKa = 10.51AKK75 pKa = 10.79LMDD78 pKa = 4.09SDD80 pKa = 4.22QRR82 pKa = 11.84CKK84 pKa = 10.15IQNSNGYY91 pKa = 9.18DD92 pKa = 3.49YY93 pKa = 11.87SHH95 pKa = 7.23IEE97 pKa = 3.98SGVLSGILKK106 pKa = 8.4TAQALVANLLTGFEE120 pKa = 4.52SHH122 pKa = 7.27FLNDD126 pKa = 3.53CSFSNGASQGFKK138 pKa = 10.78LRR140 pKa = 11.84DD141 pKa = 3.12AAPFKK146 pKa = 10.75KK147 pKa = 9.93IAGQATVTAPAYY159 pKa = 9.74DD160 pKa = 3.34IAVAAVKK167 pKa = 9.25TCAPWYY173 pKa = 10.42AYY175 pKa = 8.05MQEE178 pKa = 4.3TYY180 pKa = 11.25GDD182 pKa = 3.41EE183 pKa = 4.55TKK185 pKa = 10.08WFRR188 pKa = 11.84RR189 pKa = 11.84VYY191 pKa = 10.56GNGLFSVPKK200 pKa = 9.87NNKK203 pKa = 8.33IDD205 pKa = 3.54RR206 pKa = 11.84AACKK210 pKa = 10.1EE211 pKa = 3.91PDD213 pKa = 3.19MNMYY217 pKa = 9.01LQKK220 pKa = 10.87GAGSFIRR227 pKa = 11.84KK228 pKa = 9.06RR229 pKa = 11.84LRR231 pKa = 11.84SVGIDD236 pKa = 3.74LNDD239 pKa = 3.19QTRR242 pKa = 11.84NQEE245 pKa = 4.02LARR248 pKa = 11.84LGSIDD253 pKa = 4.18GSLATIDD260 pKa = 4.48LSSASDD266 pKa = 3.95SISDD270 pKa = 3.63RR271 pKa = 11.84LVWDD275 pKa = 4.22LLPPHH280 pKa = 6.96VYY282 pKa = 10.32SYY284 pKa = 10.91LARR287 pKa = 11.84IRR289 pKa = 11.84TSFTMIDD296 pKa = 3.38GRR298 pKa = 11.84LHH300 pKa = 5.44KK301 pKa = 9.96WGLFSTMGNGFTFEE315 pKa = 4.45LEE317 pKa = 4.66SMIFWALSKK326 pKa = 10.89SIMLSMGVTGSLGIYY341 pKa = 9.83GDD343 pKa = 4.61DD344 pKa = 3.23IIVPVEE350 pKa = 3.98CRR352 pKa = 11.84PTLLKK357 pKa = 10.79VLSAVNFLPNEE368 pKa = 4.22EE369 pKa = 4.26KK370 pKa = 10.34TFTTGYY376 pKa = 10.01FRR378 pKa = 11.84EE379 pKa = 4.32SCGAHH384 pKa = 5.96FFKK387 pKa = 11.03DD388 pKa = 3.04ADD390 pKa = 3.79MKK392 pKa = 10.79PFYY395 pKa = 10.06CKK397 pKa = 10.27RR398 pKa = 11.84PMEE401 pKa = 4.28TLPDD405 pKa = 3.68VMLLCNRR412 pKa = 11.84IRR414 pKa = 11.84GWQTVGGMSDD424 pKa = 3.34PRR426 pKa = 11.84LFPIWKK432 pKa = 8.89EE433 pKa = 3.85FADD436 pKa = 4.28MIPPKK441 pKa = 10.62FKK443 pKa = 10.78GGCNLDD449 pKa = 3.23RR450 pKa = 11.84DD451 pKa = 4.59TYY453 pKa = 11.03LVSPDD458 pKa = 3.49KK459 pKa = 11.07PGVSLVRR466 pKa = 11.84IAKK469 pKa = 9.1VRR471 pKa = 11.84SGFNHH476 pKa = 6.91AFPYY480 pKa = 9.19GHH482 pKa = 6.61EE483 pKa = 4.13NGRR486 pKa = 11.84YY487 pKa = 6.63VHH489 pKa = 6.72WLHH492 pKa = 6.32MGSGEE497 pKa = 4.1VLEE500 pKa = 5.22TISSARR506 pKa = 11.84YY507 pKa = 7.61RR508 pKa = 11.84CKK510 pKa = 10.44PNSEE514 pKa = 3.77WRR516 pKa = 11.84TQIPLFPQEE525 pKa = 5.23LEE527 pKa = 4.08ACVLSS532 pKa = 3.99
MM1 pKa = 7.12FRR3 pKa = 11.84FRR5 pKa = 11.84EE6 pKa = 4.05IEE8 pKa = 3.77KK9 pKa = 8.89TLCMDD14 pKa = 4.01RR15 pKa = 11.84TRR17 pKa = 11.84DD18 pKa = 3.47CAVRR22 pKa = 11.84FHH24 pKa = 7.27VYY26 pKa = 9.84LQSLDD31 pKa = 4.29LGSSDD36 pKa = 4.25PLSPDD41 pKa = 3.21FDD43 pKa = 3.73GLAYY47 pKa = 10.7LRR49 pKa = 11.84DD50 pKa = 3.68EE51 pKa = 5.36CLTKK55 pKa = 10.59HH56 pKa = 6.39PSLGDD61 pKa = 3.7SNSDD65 pKa = 3.12ARR67 pKa = 11.84RR68 pKa = 11.84KK69 pKa = 9.05EE70 pKa = 3.55LAYY73 pKa = 10.51AKK75 pKa = 10.79LMDD78 pKa = 4.09SDD80 pKa = 4.22QRR82 pKa = 11.84CKK84 pKa = 10.15IQNSNGYY91 pKa = 9.18DD92 pKa = 3.49YY93 pKa = 11.87SHH95 pKa = 7.23IEE97 pKa = 3.98SGVLSGILKK106 pKa = 8.4TAQALVANLLTGFEE120 pKa = 4.52SHH122 pKa = 7.27FLNDD126 pKa = 3.53CSFSNGASQGFKK138 pKa = 10.78LRR140 pKa = 11.84DD141 pKa = 3.12AAPFKK146 pKa = 10.75KK147 pKa = 9.93IAGQATVTAPAYY159 pKa = 9.74DD160 pKa = 3.34IAVAAVKK167 pKa = 9.25TCAPWYY173 pKa = 10.42AYY175 pKa = 8.05MQEE178 pKa = 4.3TYY180 pKa = 11.25GDD182 pKa = 3.41EE183 pKa = 4.55TKK185 pKa = 10.08WFRR188 pKa = 11.84RR189 pKa = 11.84VYY191 pKa = 10.56GNGLFSVPKK200 pKa = 9.87NNKK203 pKa = 8.33IDD205 pKa = 3.54RR206 pKa = 11.84AACKK210 pKa = 10.1EE211 pKa = 3.91PDD213 pKa = 3.19MNMYY217 pKa = 9.01LQKK220 pKa = 10.87GAGSFIRR227 pKa = 11.84KK228 pKa = 9.06RR229 pKa = 11.84LRR231 pKa = 11.84SVGIDD236 pKa = 3.74LNDD239 pKa = 3.19QTRR242 pKa = 11.84NQEE245 pKa = 4.02LARR248 pKa = 11.84LGSIDD253 pKa = 4.18GSLATIDD260 pKa = 4.48LSSASDD266 pKa = 3.95SISDD270 pKa = 3.63RR271 pKa = 11.84LVWDD275 pKa = 4.22LLPPHH280 pKa = 6.96VYY282 pKa = 10.32SYY284 pKa = 10.91LARR287 pKa = 11.84IRR289 pKa = 11.84TSFTMIDD296 pKa = 3.38GRR298 pKa = 11.84LHH300 pKa = 5.44KK301 pKa = 9.96WGLFSTMGNGFTFEE315 pKa = 4.45LEE317 pKa = 4.66SMIFWALSKK326 pKa = 10.89SIMLSMGVTGSLGIYY341 pKa = 9.83GDD343 pKa = 4.61DD344 pKa = 3.23IIVPVEE350 pKa = 3.98CRR352 pKa = 11.84PTLLKK357 pKa = 10.79VLSAVNFLPNEE368 pKa = 4.22EE369 pKa = 4.26KK370 pKa = 10.34TFTTGYY376 pKa = 10.01FRR378 pKa = 11.84EE379 pKa = 4.32SCGAHH384 pKa = 5.96FFKK387 pKa = 11.03DD388 pKa = 3.04ADD390 pKa = 3.79MKK392 pKa = 10.79PFYY395 pKa = 10.06CKK397 pKa = 10.27RR398 pKa = 11.84PMEE401 pKa = 4.28TLPDD405 pKa = 3.68VMLLCNRR412 pKa = 11.84IRR414 pKa = 11.84GWQTVGGMSDD424 pKa = 3.34PRR426 pKa = 11.84LFPIWKK432 pKa = 8.89EE433 pKa = 3.85FADD436 pKa = 4.28MIPPKK441 pKa = 10.62FKK443 pKa = 10.78GGCNLDD449 pKa = 3.23RR450 pKa = 11.84DD451 pKa = 4.59TYY453 pKa = 11.03LVSPDD458 pKa = 3.49KK459 pKa = 11.07PGVSLVRR466 pKa = 11.84IAKK469 pKa = 9.1VRR471 pKa = 11.84SGFNHH476 pKa = 6.91AFPYY480 pKa = 9.19GHH482 pKa = 6.61EE483 pKa = 4.13NGRR486 pKa = 11.84YY487 pKa = 6.63VHH489 pKa = 6.72WLHH492 pKa = 6.32MGSGEE497 pKa = 4.1VLEE500 pKa = 5.22TISSARR506 pKa = 11.84YY507 pKa = 7.61RR508 pKa = 11.84CKK510 pKa = 10.44PNSEE514 pKa = 3.77WRR516 pKa = 11.84TQIPLFPQEE525 pKa = 5.23LEE527 pKa = 4.08ACVLSS532 pKa = 3.99
Molecular weight: 59.93 kDa
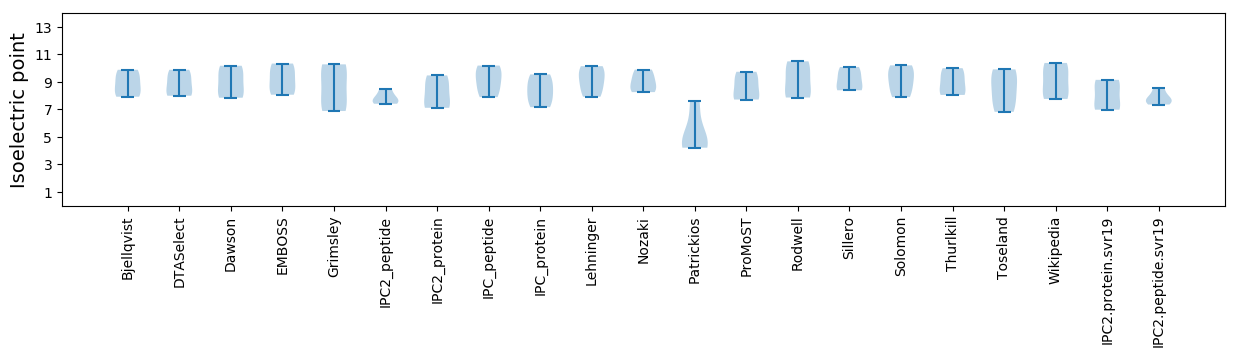
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P07394|MATA_BPGA Maturation protein A OS=Enterobacteria phage GA OX=12018 GN=A PE=2 SV=1
MM1 pKa = 7.25FPKK4 pKa = 10.75SNIDD8 pKa = 3.01RR9 pKa = 11.84NYY11 pKa = 9.11KK12 pKa = 9.97VKK14 pKa = 10.83LISYY18 pKa = 7.89DD19 pKa = 3.35KK20 pKa = 10.7KK21 pKa = 11.21GKK23 pKa = 9.86LVSDD27 pKa = 4.28DD28 pKa = 4.13SFEE31 pKa = 4.16QVEE34 pKa = 4.6NYY36 pKa = 10.19LFQNRR41 pKa = 11.84STTYY45 pKa = 8.33KK46 pKa = 9.31PGYY49 pKa = 8.38IRR51 pKa = 11.84RR52 pKa = 11.84DD53 pKa = 3.36FRR55 pKa = 11.84RR56 pKa = 11.84PTNFWNGYY64 pKa = 8.42RR65 pKa = 11.84CFNQPVGTFTRR76 pKa = 11.84KK77 pKa = 10.07LSDD80 pKa = 3.08GGRR83 pKa = 11.84QVADD87 pKa = 3.47YY88 pKa = 11.18GIVNPNKK95 pKa = 8.75FTANSQHH102 pKa = 6.72LGDD105 pKa = 4.56NMVIYY110 pKa = 9.42PGPFSINIDD119 pKa = 3.04QRR121 pKa = 11.84ASVEE125 pKa = 4.17VLNKK129 pKa = 10.41LSQSNLNIGVAIAEE143 pKa = 4.38AKK145 pKa = 7.55MTASLLAKK153 pKa = 10.16QSIALIRR160 pKa = 11.84AYY162 pKa = 9.19TAAKK166 pKa = 8.83RR167 pKa = 11.84GNWRR171 pKa = 11.84EE172 pKa = 3.86VLSQLLISEE181 pKa = 4.29HH182 pKa = 6.74RR183 pKa = 11.84FRR185 pKa = 11.84APAKK189 pKa = 10.09DD190 pKa = 3.66LGGRR194 pKa = 11.84WLEE197 pKa = 4.16LQYY200 pKa = 11.47GWLPLMSDD208 pKa = 3.94LKK210 pKa = 10.95AAYY213 pKa = 9.66DD214 pKa = 4.15LLTQTKK220 pKa = 9.72LPAFMPLRR228 pKa = 11.84VTRR231 pKa = 11.84TVGGTHH237 pKa = 6.26NYY239 pKa = 9.16KK240 pKa = 9.95VRR242 pKa = 11.84NVEE245 pKa = 4.16SAGDD249 pKa = 3.41TWSYY253 pKa = 9.95RR254 pKa = 11.84HH255 pKa = 6.22RR256 pKa = 11.84LSVNYY261 pKa = 9.63RR262 pKa = 11.84IWYY265 pKa = 7.99FISDD269 pKa = 4.2PRR271 pKa = 11.84LAWASSLGLLNPLEE285 pKa = 4.69IYY287 pKa = 9.32WEE289 pKa = 4.0KK290 pKa = 10.17TPWSFVVDD298 pKa = 3.47WFLPVGNLIEE308 pKa = 5.13AMSNPLGLDD317 pKa = 3.5IISGTKK323 pKa = 7.87TWQLEE328 pKa = 4.42SKK330 pKa = 10.88LNATLPASGWSGTAKK345 pKa = 9.2LTAYY349 pKa = 10.14AKK351 pKa = 10.52AYY353 pKa = 10.06DD354 pKa = 3.54RR355 pKa = 11.84STFYY359 pKa = 11.1SFPTPLPYY367 pKa = 10.52VKK369 pKa = 10.53SPLSGLHH376 pKa = 6.17LANALALINQRR387 pKa = 11.84LKK389 pKa = 10.91RR390 pKa = 3.85
MM1 pKa = 7.25FPKK4 pKa = 10.75SNIDD8 pKa = 3.01RR9 pKa = 11.84NYY11 pKa = 9.11KK12 pKa = 9.97VKK14 pKa = 10.83LISYY18 pKa = 7.89DD19 pKa = 3.35KK20 pKa = 10.7KK21 pKa = 11.21GKK23 pKa = 9.86LVSDD27 pKa = 4.28DD28 pKa = 4.13SFEE31 pKa = 4.16QVEE34 pKa = 4.6NYY36 pKa = 10.19LFQNRR41 pKa = 11.84STTYY45 pKa = 8.33KK46 pKa = 9.31PGYY49 pKa = 8.38IRR51 pKa = 11.84RR52 pKa = 11.84DD53 pKa = 3.36FRR55 pKa = 11.84RR56 pKa = 11.84PTNFWNGYY64 pKa = 8.42RR65 pKa = 11.84CFNQPVGTFTRR76 pKa = 11.84KK77 pKa = 10.07LSDD80 pKa = 3.08GGRR83 pKa = 11.84QVADD87 pKa = 3.47YY88 pKa = 11.18GIVNPNKK95 pKa = 8.75FTANSQHH102 pKa = 6.72LGDD105 pKa = 4.56NMVIYY110 pKa = 9.42PGPFSINIDD119 pKa = 3.04QRR121 pKa = 11.84ASVEE125 pKa = 4.17VLNKK129 pKa = 10.41LSQSNLNIGVAIAEE143 pKa = 4.38AKK145 pKa = 7.55MTASLLAKK153 pKa = 10.16QSIALIRR160 pKa = 11.84AYY162 pKa = 9.19TAAKK166 pKa = 8.83RR167 pKa = 11.84GNWRR171 pKa = 11.84EE172 pKa = 3.86VLSQLLISEE181 pKa = 4.29HH182 pKa = 6.74RR183 pKa = 11.84FRR185 pKa = 11.84APAKK189 pKa = 10.09DD190 pKa = 3.66LGGRR194 pKa = 11.84WLEE197 pKa = 4.16LQYY200 pKa = 11.47GWLPLMSDD208 pKa = 3.94LKK210 pKa = 10.95AAYY213 pKa = 9.66DD214 pKa = 4.15LLTQTKK220 pKa = 9.72LPAFMPLRR228 pKa = 11.84VTRR231 pKa = 11.84TVGGTHH237 pKa = 6.26NYY239 pKa = 9.16KK240 pKa = 9.95VRR242 pKa = 11.84NVEE245 pKa = 4.16SAGDD249 pKa = 3.41TWSYY253 pKa = 9.95RR254 pKa = 11.84HH255 pKa = 6.22RR256 pKa = 11.84LSVNYY261 pKa = 9.63RR262 pKa = 11.84IWYY265 pKa = 7.99FISDD269 pKa = 4.2PRR271 pKa = 11.84LAWASSLGLLNPLEE285 pKa = 4.69IYY287 pKa = 9.32WEE289 pKa = 4.0KK290 pKa = 10.17TPWSFVVDD298 pKa = 3.47WFLPVGNLIEE308 pKa = 5.13AMSNPLGLDD317 pKa = 3.5IISGTKK323 pKa = 7.87TWQLEE328 pKa = 4.42SKK330 pKa = 10.88LNATLPASGWSGTAKK345 pKa = 9.2LTAYY349 pKa = 10.14AKK351 pKa = 10.52AYY353 pKa = 10.06DD354 pKa = 3.54RR355 pKa = 11.84STFYY359 pKa = 11.1SFPTPLPYY367 pKa = 10.52VKK369 pKa = 10.53SPLSGLHH376 pKa = 6.17LANALALINQRR387 pKa = 11.84LKK389 pKa = 10.91RR390 pKa = 3.85
Molecular weight: 44.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1115 |
63 |
532 |
278.8 |
31.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.713 ± 1.191 | 1.435 ± 0.743 |
5.471 ± 0.808 | 3.767 ± 0.458 |
4.664 ± 0.53 | 6.726 ± 0.494 |
1.614 ± 0.383 | 5.112 ± 0.522 |
5.74 ± 0.225 | 10.404 ± 1.075 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.422 ± 0.551 | 4.933 ± 0.775 |
4.753 ± 0.549 | 2.691 ± 0.328 |
6.099 ± 0.811 | 8.7 ± 0.592 |
5.291 ± 0.273 | 5.919 ± 1.637 |
2.332 ± 0.48 | 4.215 ± 0.442 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
