
Berrimah virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Ephemerovirus; Berrimah ephemerovirus
Average proteome isoelectric point is 7.02
Get precalculated fractions of proteins
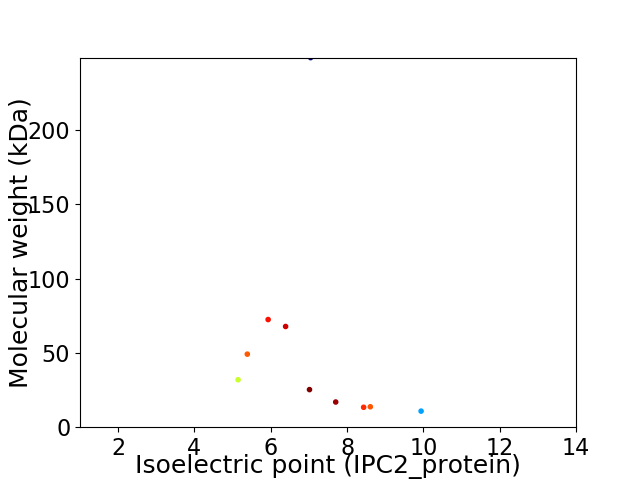
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
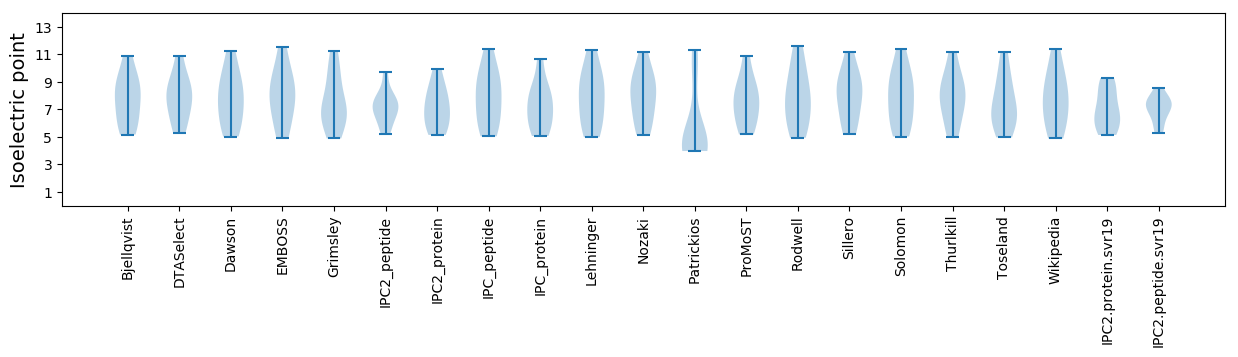
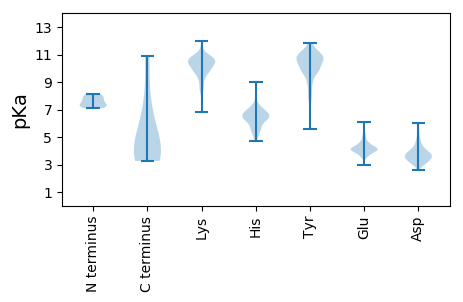
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I3NUX6|I3NUX6_9RHAB Matrix protein M OS=Berrimah virus OX=318834 GN=M PE=4 SV=1
MM1 pKa = 7.25SHH3 pKa = 6.33QKK5 pKa = 9.93VKK7 pKa = 10.89LMTGTYY13 pKa = 10.29DD14 pKa = 3.34AGKK17 pKa = 9.36LQRR20 pKa = 11.84NLQEE24 pKa = 5.08HH25 pKa = 6.58ILLEE29 pKa = 4.24EE30 pKa = 4.9DD31 pKa = 3.47EE32 pKa = 5.58DD33 pKa = 4.88GSFAQGDD40 pKa = 3.82QVAEE44 pKa = 4.6EE45 pKa = 4.32ILPINPLLNKK55 pKa = 9.91SEE57 pKa = 4.06PKK59 pKa = 10.34VYY61 pKa = 10.0PIEE64 pKa = 4.68INLEE68 pKa = 4.08DD69 pKa = 4.0LEE71 pKa = 4.38KK72 pKa = 11.33ANGIEE77 pKa = 4.57DD78 pKa = 3.83DD79 pKa = 4.21WEE81 pKa = 4.77HH82 pKa = 6.5SLLKK86 pKa = 10.12IIEE89 pKa = 4.24SSEE92 pKa = 3.98VSPKK96 pKa = 10.72LSWDD100 pKa = 3.85DD101 pKa = 3.68EE102 pKa = 4.73FEE104 pKa = 4.33SDD106 pKa = 5.42SYY108 pKa = 11.87EE109 pKa = 3.99KK110 pKa = 10.5DD111 pKa = 3.13KK112 pKa = 11.62KK113 pKa = 10.9GMRR116 pKa = 11.84GPKK119 pKa = 9.18QMKK122 pKa = 9.92DD123 pKa = 2.96IQNKK127 pKa = 9.16YY128 pKa = 9.31IVEE131 pKa = 4.01IDD133 pKa = 3.49EE134 pKa = 5.78DD135 pKa = 4.02SLKK138 pKa = 11.14DD139 pKa = 3.6VLKK142 pKa = 10.78VLSAFEE148 pKa = 3.9IRR150 pKa = 11.84EE151 pKa = 4.19GEE153 pKa = 4.17DD154 pKa = 3.38FNVSNNKK161 pKa = 9.29SKK163 pKa = 10.57RR164 pKa = 11.84VKK166 pKa = 10.15IVIEE170 pKa = 3.79KK171 pKa = 10.53RR172 pKa = 11.84GTRR175 pKa = 11.84EE176 pKa = 3.73CNKK179 pKa = 9.68IDD181 pKa = 3.64VTGKK185 pKa = 9.98SPQNEE190 pKa = 3.74AMKK193 pKa = 10.37EE194 pKa = 3.88DD195 pKa = 3.65RR196 pKa = 11.84SKK198 pKa = 11.15FDD200 pKa = 3.44QIMDD204 pKa = 3.31QFQRR208 pKa = 11.84GIRR211 pKa = 11.84IKK213 pKa = 10.66KK214 pKa = 9.86KK215 pKa = 9.63FGKK218 pKa = 10.57GYY220 pKa = 11.25VKK222 pKa = 10.33MNADD226 pKa = 3.77NMPGTYY232 pKa = 9.65HH233 pKa = 6.86EE234 pKa = 5.24LYY236 pKa = 10.28SVVSGIVDD244 pKa = 3.59EE245 pKa = 4.56NTTEE249 pKa = 5.3DD250 pKa = 3.4MIKK253 pKa = 10.61YY254 pKa = 9.66LFRR257 pKa = 11.84KK258 pKa = 9.5SKK260 pKa = 10.47SFKK263 pKa = 11.25AMGKK267 pKa = 8.32MLNIDD272 pKa = 4.51EE273 pKa = 5.76IILCC277 pKa = 4.54
MM1 pKa = 7.25SHH3 pKa = 6.33QKK5 pKa = 9.93VKK7 pKa = 10.89LMTGTYY13 pKa = 10.29DD14 pKa = 3.34AGKK17 pKa = 9.36LQRR20 pKa = 11.84NLQEE24 pKa = 5.08HH25 pKa = 6.58ILLEE29 pKa = 4.24EE30 pKa = 4.9DD31 pKa = 3.47EE32 pKa = 5.58DD33 pKa = 4.88GSFAQGDD40 pKa = 3.82QVAEE44 pKa = 4.6EE45 pKa = 4.32ILPINPLLNKK55 pKa = 9.91SEE57 pKa = 4.06PKK59 pKa = 10.34VYY61 pKa = 10.0PIEE64 pKa = 4.68INLEE68 pKa = 4.08DD69 pKa = 4.0LEE71 pKa = 4.38KK72 pKa = 11.33ANGIEE77 pKa = 4.57DD78 pKa = 3.83DD79 pKa = 4.21WEE81 pKa = 4.77HH82 pKa = 6.5SLLKK86 pKa = 10.12IIEE89 pKa = 4.24SSEE92 pKa = 3.98VSPKK96 pKa = 10.72LSWDD100 pKa = 3.85DD101 pKa = 3.68EE102 pKa = 4.73FEE104 pKa = 4.33SDD106 pKa = 5.42SYY108 pKa = 11.87EE109 pKa = 3.99KK110 pKa = 10.5DD111 pKa = 3.13KK112 pKa = 11.62KK113 pKa = 10.9GMRR116 pKa = 11.84GPKK119 pKa = 9.18QMKK122 pKa = 9.92DD123 pKa = 2.96IQNKK127 pKa = 9.16YY128 pKa = 9.31IVEE131 pKa = 4.01IDD133 pKa = 3.49EE134 pKa = 5.78DD135 pKa = 4.02SLKK138 pKa = 11.14DD139 pKa = 3.6VLKK142 pKa = 10.78VLSAFEE148 pKa = 3.9IRR150 pKa = 11.84EE151 pKa = 4.19GEE153 pKa = 4.17DD154 pKa = 3.38FNVSNNKK161 pKa = 9.29SKK163 pKa = 10.57RR164 pKa = 11.84VKK166 pKa = 10.15IVIEE170 pKa = 3.79KK171 pKa = 10.53RR172 pKa = 11.84GTRR175 pKa = 11.84EE176 pKa = 3.73CNKK179 pKa = 9.68IDD181 pKa = 3.64VTGKK185 pKa = 9.98SPQNEE190 pKa = 3.74AMKK193 pKa = 10.37EE194 pKa = 3.88DD195 pKa = 3.65RR196 pKa = 11.84SKK198 pKa = 11.15FDD200 pKa = 3.44QIMDD204 pKa = 3.31QFQRR208 pKa = 11.84GIRR211 pKa = 11.84IKK213 pKa = 10.66KK214 pKa = 9.86KK215 pKa = 9.63FGKK218 pKa = 10.57GYY220 pKa = 11.25VKK222 pKa = 10.33MNADD226 pKa = 3.77NMPGTYY232 pKa = 9.65HH233 pKa = 6.86EE234 pKa = 5.24LYY236 pKa = 10.28SVVSGIVDD244 pKa = 3.59EE245 pKa = 4.56NTTEE249 pKa = 5.3DD250 pKa = 3.4MIKK253 pKa = 10.61YY254 pKa = 9.66LFRR257 pKa = 11.84KK258 pKa = 9.5SKK260 pKa = 10.47SFKK263 pKa = 11.25AMGKK267 pKa = 8.32MLNIDD272 pKa = 4.51EE273 pKa = 5.76IILCC277 pKa = 4.54
Molecular weight: 31.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I3NUX9|I3NUX9_9RHAB Isoform of I3NUY0 Alpha 1 protein OS=Berrimah virus OX=318834 GN=alpha PE=4 SV=1
MM1 pKa = 7.91DD2 pKa = 5.77RR3 pKa = 11.84SLLSNFWKK11 pKa = 10.69DD12 pKa = 3.22FTNWSEE18 pKa = 3.61ARR20 pKa = 11.84KK21 pKa = 9.86IEE23 pKa = 5.23IIDD26 pKa = 2.88WWTNLEE32 pKa = 3.74GRR34 pKa = 11.84IRR36 pKa = 11.84LGFWIIFIILIGLIIIRR53 pKa = 11.84ISYY56 pKa = 9.9KK57 pKa = 9.84ILKK60 pKa = 9.53CIQLLRR66 pKa = 11.84KK67 pKa = 10.14GIVKK71 pKa = 8.65TKK73 pKa = 10.53RR74 pKa = 11.84IIKK77 pKa = 9.39KK78 pKa = 9.47RR79 pKa = 11.84KK80 pKa = 7.02TIKK83 pKa = 10.25KK84 pKa = 8.21YY85 pKa = 10.24RR86 pKa = 11.84KK87 pKa = 8.8HH88 pKa = 5.82
MM1 pKa = 7.91DD2 pKa = 5.77RR3 pKa = 11.84SLLSNFWKK11 pKa = 10.69DD12 pKa = 3.22FTNWSEE18 pKa = 3.61ARR20 pKa = 11.84KK21 pKa = 9.86IEE23 pKa = 5.23IIDD26 pKa = 2.88WWTNLEE32 pKa = 3.74GRR34 pKa = 11.84IRR36 pKa = 11.84LGFWIIFIILIGLIIIRR53 pKa = 11.84ISYY56 pKa = 9.9KK57 pKa = 9.84ILKK60 pKa = 9.53CIQLLRR66 pKa = 11.84KK67 pKa = 10.14GIVKK71 pKa = 8.65TKK73 pKa = 10.53RR74 pKa = 11.84IIKK77 pKa = 9.39KK78 pKa = 9.47RR79 pKa = 11.84KK80 pKa = 7.02TIKK83 pKa = 10.25KK84 pKa = 8.21YY85 pKa = 10.24RR86 pKa = 11.84KK87 pKa = 8.8HH88 pKa = 5.82
Molecular weight: 10.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4745 |
88 |
2144 |
474.5 |
55.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.929 ± 0.426 | 1.855 ± 0.268 |
5.606 ± 0.48 | 8.114 ± 0.513 |
3.983 ± 0.3 | 5.564 ± 0.274 |
2.234 ± 0.165 | 8.746 ± 0.772 |
8.809 ± 0.682 | 9.505 ± 0.803 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.655 ± 0.271 | 6.596 ± 0.418 |
3.182 ± 0.213 | 2.655 ± 0.241 |
5.016 ± 0.353 | 7.503 ± 0.494 |
4.215 ± 0.303 | 4.953 ± 0.431 |
1.812 ± 0.267 | 4.067 ± 0.378 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
