
Sarcina sp. DSM 11001
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Sarcina; unclassified Sarcina
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
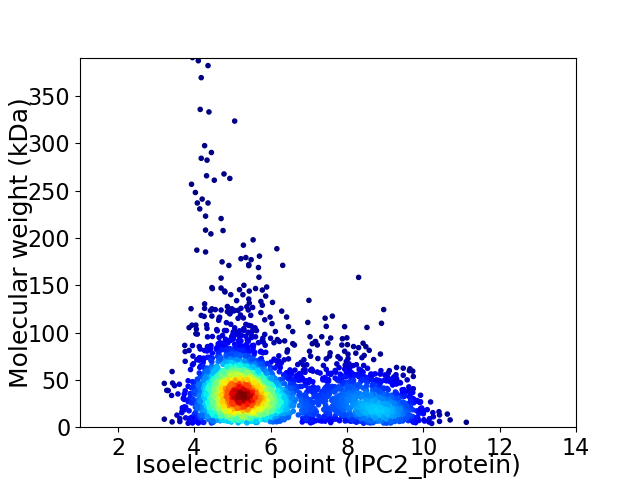
Virtual 2D-PAGE plot for 3187 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G9F2P0|A0A1G9F2P0_9CLOT TPM_phosphatase domain-containing protein OS=Sarcina sp. DSM 11001 OX=1798184 GN=SAMN04487833_1085 PE=4 SV=1
MM1 pKa = 7.39HH2 pKa = 7.73KK3 pKa = 10.14NRR5 pKa = 11.84KK6 pKa = 8.93RR7 pKa = 11.84FQALVLAAAITLAAVPGTGFTARR30 pKa = 11.84AAEE33 pKa = 4.08PEE35 pKa = 4.31IPAEE39 pKa = 3.77TAYY42 pKa = 9.88PLEE45 pKa = 4.34PEE47 pKa = 4.45IPAEE51 pKa = 3.72TAYY54 pKa = 9.88PLEE57 pKa = 4.34PEE59 pKa = 4.45IPAEE63 pKa = 3.78TAYY66 pKa = 9.94PSEE69 pKa = 4.31PEE71 pKa = 4.0IPAEE75 pKa = 3.67TAYY78 pKa = 9.98PLEE81 pKa = 4.92AEE83 pKa = 4.37EE84 pKa = 4.19TAADD88 pKa = 3.84AVDD91 pKa = 6.11GIALPAVSLNQEE103 pKa = 3.64EE104 pKa = 5.32SVFGEE109 pKa = 4.23IPAEE113 pKa = 4.04DD114 pKa = 4.76EE115 pKa = 4.23IEE117 pKa = 4.09EE118 pKa = 4.83DD119 pKa = 3.42STNPEE124 pKa = 4.35DD125 pKa = 4.55EE126 pKa = 4.59ILPEE130 pKa = 4.57GDD132 pKa = 3.84SPDD135 pKa = 3.33IQEE138 pKa = 4.97DD139 pKa = 4.12VSSEE143 pKa = 4.03PTGGTVSQEE152 pKa = 4.01NPEE155 pKa = 3.98EE156 pKa = 4.35DD157 pKa = 3.63SFPEE161 pKa = 3.98NPEE164 pKa = 3.74EE165 pKa = 5.09KK166 pKa = 10.51GAAEE170 pKa = 5.4NPDD173 pKa = 2.79QDD175 pKa = 3.73AASEE179 pKa = 4.23ITEE182 pKa = 4.18EE183 pKa = 4.11TDD185 pKa = 3.24PAEE188 pKa = 4.66TPDD191 pKa = 3.63QDD193 pKa = 3.83VASEE197 pKa = 4.04NLEE200 pKa = 3.9EE201 pKa = 4.04TDD203 pKa = 3.16AQNFPEE209 pKa = 4.57EE210 pKa = 4.35NTSEE214 pKa = 4.2EE215 pKa = 4.29PAEE218 pKa = 4.36GAAQEE223 pKa = 4.65TEE225 pKa = 4.28TVTQSVDD232 pKa = 3.13EE233 pKa = 4.31VRR235 pKa = 11.84EE236 pKa = 4.09KK237 pKa = 10.65TDD239 pKa = 4.11VEE241 pKa = 4.84HH242 pKa = 7.38EE243 pKa = 4.4DD244 pKa = 3.3NDD246 pKa = 4.56LLFEE250 pKa = 4.66EE251 pKa = 4.87YY252 pKa = 10.85AEE254 pKa = 4.57AMFEE258 pKa = 4.03EE259 pKa = 5.09AADD262 pKa = 4.13GAQPGTSGRR271 pKa = 11.84GQKK274 pKa = 8.24NTGDD278 pKa = 3.44QLTGQDD284 pKa = 3.09RR285 pKa = 11.84VIYY288 pKa = 10.02DD289 pKa = 3.65ALKK292 pKa = 10.53AAALEE297 pKa = 4.3IADD300 pKa = 4.17GQRR303 pKa = 11.84DD304 pKa = 3.78STIVEE309 pKa = 4.13IPLSEE314 pKa = 4.4LGIDD318 pKa = 3.75AGKK321 pKa = 10.57QYY323 pKa = 9.58TAEE326 pKa = 4.67DD327 pKa = 3.64LGLDD331 pKa = 3.58YY332 pKa = 10.49IYY334 pKa = 10.92RR335 pKa = 11.84RR336 pKa = 11.84VNGTGEE342 pKa = 4.13WNPAIEE348 pKa = 4.39DD349 pKa = 3.86AVNSMLSFDD358 pKa = 3.29SRR360 pKa = 11.84EE361 pKa = 3.53IFRR364 pKa = 11.84RR365 pKa = 11.84LWADD369 pKa = 3.6CPYY372 pKa = 11.21EE373 pKa = 4.15MYY375 pKa = 8.71WQKK378 pKa = 11.07GGYY381 pKa = 8.69SFPVSLGCSKK391 pKa = 10.51RR392 pKa = 11.84ASGSGDD398 pKa = 2.8SWSGFVVIEE407 pKa = 3.93EE408 pKa = 4.52SPVTFSMCVEE418 pKa = 3.64QKK420 pKa = 10.49YY421 pKa = 9.03RR422 pKa = 11.84QDD424 pKa = 3.23QDD426 pKa = 4.28DD427 pKa = 4.76EE428 pKa = 5.37YY429 pKa = 11.78SVDD432 pKa = 3.49TSKK435 pKa = 9.75TGAACSAALFARR447 pKa = 11.84SIVNDD452 pKa = 3.72ADD454 pKa = 3.54NSGLSDD460 pKa = 3.76YY461 pKa = 11.43DD462 pKa = 3.3RR463 pKa = 11.84LVYY466 pKa = 10.79YY467 pKa = 10.41KK468 pKa = 10.72DD469 pKa = 4.03RR470 pKa = 11.84ICNEE474 pKa = 3.43VVYY477 pKa = 10.99NEE479 pKa = 4.2EE480 pKa = 3.76ARR482 pKa = 11.84QNSEE486 pKa = 4.26TYY488 pKa = 9.87PDD490 pKa = 3.71RR491 pKa = 11.84GPWALIYY498 pKa = 11.08VFDD501 pKa = 4.52QNPDD505 pKa = 3.43TNVVCEE511 pKa = 4.79GYY513 pKa = 10.22SEE515 pKa = 4.36AFQYY519 pKa = 11.24LCGLTEE525 pKa = 4.02FQSRR529 pKa = 11.84KK530 pKa = 7.39IQVYY534 pKa = 9.47SVSGMFGGGTGAGPHH549 pKa = 6.23KK550 pKa = 10.27WNIVHH555 pKa = 6.79MDD557 pKa = 3.54DD558 pKa = 4.39GLNYY562 pKa = 10.45LADD565 pKa = 3.64VTNSDD570 pKa = 4.15EE571 pKa = 4.66GTVGWDD577 pKa = 2.89GRR579 pKa = 11.84LFLKK583 pKa = 10.79GIEE586 pKa = 4.1GNVYY590 pKa = 10.49DD591 pKa = 5.06GYY593 pKa = 9.89VLSWEE598 pKa = 4.47EE599 pKa = 3.7YY600 pKa = 7.98QEE602 pKa = 4.13EE603 pKa = 4.49VEE605 pKa = 4.91EE606 pKa = 4.09EE607 pKa = 4.18DD608 pKa = 3.04GTYY611 pKa = 10.79VYY613 pKa = 9.21TYY615 pKa = 9.64PAGSVDD621 pKa = 3.48YY622 pKa = 10.67AYY624 pKa = 10.53DD625 pKa = 3.37QEE627 pKa = 4.25TRR629 pKa = 11.84ALFTDD634 pKa = 3.82EE635 pKa = 5.13EE636 pKa = 4.71LTLSSGDD643 pKa = 3.87YY644 pKa = 11.04VPGSLPPGPPAPTVDD659 pKa = 3.99EE660 pKa = 4.11IVSIEE665 pKa = 4.11EE666 pKa = 4.05VYY668 pKa = 10.76LYY670 pKa = 9.75LTDD673 pKa = 4.58KK674 pKa = 11.18IEE676 pKa = 3.97MRR678 pKa = 11.84FCAKK682 pKa = 10.37VEE684 pKa = 4.21GNIEE688 pKa = 3.76QDD690 pKa = 3.29DD691 pKa = 5.01YY692 pKa = 11.58ITFICAGRR700 pKa = 11.84TVTQTVSEE708 pKa = 4.68AEE710 pKa = 4.01TDD712 pKa = 3.57SEE714 pKa = 4.23GRR716 pKa = 11.84LVFSLEE722 pKa = 3.52LAARR726 pKa = 11.84QMTDD730 pKa = 2.32QVTYY734 pKa = 10.76FMTVDD739 pKa = 3.94GTAGTSGQYY748 pKa = 9.3SVRR751 pKa = 11.84SYY753 pKa = 11.94ADD755 pKa = 3.32TILNQSGMRR764 pKa = 11.84DD765 pKa = 3.45YY766 pKa = 11.33KK767 pKa = 11.08GLLRR771 pKa = 11.84AMLNYY776 pKa = 10.39GSYY779 pKa = 7.79TQLYY783 pKa = 8.47AGYY786 pKa = 8.05RR787 pKa = 11.84TDD789 pKa = 4.57FLAAEE794 pKa = 4.35GLYY797 pKa = 10.02TDD799 pKa = 3.92EE800 pKa = 4.73TDD802 pKa = 3.38PVLNMEE808 pKa = 4.96GPDD811 pKa = 3.67LTDD814 pKa = 3.85FAYY817 pKa = 9.42TYY819 pKa = 10.97KK820 pKa = 10.83LNSKK824 pKa = 9.1TDD826 pKa = 3.01GLTIGKK832 pKa = 10.1ASLLLGTDD840 pKa = 3.91LSLRR844 pKa = 11.84LYY846 pKa = 10.36YY847 pKa = 10.47EE848 pKa = 4.32PGEE851 pKa = 4.35GKK853 pKa = 8.54TDD855 pKa = 3.11KK856 pKa = 10.95SYY858 pKa = 11.4SIVLADD864 pKa = 4.08RR865 pKa = 11.84SGLEE869 pKa = 3.93PVLGYY874 pKa = 10.96DD875 pKa = 3.89DD876 pKa = 4.9TIGMYY881 pKa = 10.33YY882 pKa = 10.81ADD884 pKa = 4.88IEE886 pKa = 5.19HH887 pKa = 6.71ITPTQIGDD895 pKa = 3.47MFKK898 pKa = 11.01LDD900 pKa = 4.94FYY902 pKa = 11.82AEE904 pKa = 4.6DD905 pKa = 4.0GQAADD910 pKa = 4.07TPAASVTFGPLSYY923 pKa = 10.64CKK925 pKa = 10.32GVLEE929 pKa = 4.95SGSSSDD935 pKa = 4.07EE936 pKa = 3.95LVNLCRR942 pKa = 11.84ALYY945 pKa = 9.31EE946 pKa = 3.9YY947 pKa = 9.5WQIAIRR953 pKa = 11.84PLTT956 pKa = 3.64
MM1 pKa = 7.39HH2 pKa = 7.73KK3 pKa = 10.14NRR5 pKa = 11.84KK6 pKa = 8.93RR7 pKa = 11.84FQALVLAAAITLAAVPGTGFTARR30 pKa = 11.84AAEE33 pKa = 4.08PEE35 pKa = 4.31IPAEE39 pKa = 3.77TAYY42 pKa = 9.88PLEE45 pKa = 4.34PEE47 pKa = 4.45IPAEE51 pKa = 3.72TAYY54 pKa = 9.88PLEE57 pKa = 4.34PEE59 pKa = 4.45IPAEE63 pKa = 3.78TAYY66 pKa = 9.94PSEE69 pKa = 4.31PEE71 pKa = 4.0IPAEE75 pKa = 3.67TAYY78 pKa = 9.98PLEE81 pKa = 4.92AEE83 pKa = 4.37EE84 pKa = 4.19TAADD88 pKa = 3.84AVDD91 pKa = 6.11GIALPAVSLNQEE103 pKa = 3.64EE104 pKa = 5.32SVFGEE109 pKa = 4.23IPAEE113 pKa = 4.04DD114 pKa = 4.76EE115 pKa = 4.23IEE117 pKa = 4.09EE118 pKa = 4.83DD119 pKa = 3.42STNPEE124 pKa = 4.35DD125 pKa = 4.55EE126 pKa = 4.59ILPEE130 pKa = 4.57GDD132 pKa = 3.84SPDD135 pKa = 3.33IQEE138 pKa = 4.97DD139 pKa = 4.12VSSEE143 pKa = 4.03PTGGTVSQEE152 pKa = 4.01NPEE155 pKa = 3.98EE156 pKa = 4.35DD157 pKa = 3.63SFPEE161 pKa = 3.98NPEE164 pKa = 3.74EE165 pKa = 5.09KK166 pKa = 10.51GAAEE170 pKa = 5.4NPDD173 pKa = 2.79QDD175 pKa = 3.73AASEE179 pKa = 4.23ITEE182 pKa = 4.18EE183 pKa = 4.11TDD185 pKa = 3.24PAEE188 pKa = 4.66TPDD191 pKa = 3.63QDD193 pKa = 3.83VASEE197 pKa = 4.04NLEE200 pKa = 3.9EE201 pKa = 4.04TDD203 pKa = 3.16AQNFPEE209 pKa = 4.57EE210 pKa = 4.35NTSEE214 pKa = 4.2EE215 pKa = 4.29PAEE218 pKa = 4.36GAAQEE223 pKa = 4.65TEE225 pKa = 4.28TVTQSVDD232 pKa = 3.13EE233 pKa = 4.31VRR235 pKa = 11.84EE236 pKa = 4.09KK237 pKa = 10.65TDD239 pKa = 4.11VEE241 pKa = 4.84HH242 pKa = 7.38EE243 pKa = 4.4DD244 pKa = 3.3NDD246 pKa = 4.56LLFEE250 pKa = 4.66EE251 pKa = 4.87YY252 pKa = 10.85AEE254 pKa = 4.57AMFEE258 pKa = 4.03EE259 pKa = 5.09AADD262 pKa = 4.13GAQPGTSGRR271 pKa = 11.84GQKK274 pKa = 8.24NTGDD278 pKa = 3.44QLTGQDD284 pKa = 3.09RR285 pKa = 11.84VIYY288 pKa = 10.02DD289 pKa = 3.65ALKK292 pKa = 10.53AAALEE297 pKa = 4.3IADD300 pKa = 4.17GQRR303 pKa = 11.84DD304 pKa = 3.78STIVEE309 pKa = 4.13IPLSEE314 pKa = 4.4LGIDD318 pKa = 3.75AGKK321 pKa = 10.57QYY323 pKa = 9.58TAEE326 pKa = 4.67DD327 pKa = 3.64LGLDD331 pKa = 3.58YY332 pKa = 10.49IYY334 pKa = 10.92RR335 pKa = 11.84RR336 pKa = 11.84VNGTGEE342 pKa = 4.13WNPAIEE348 pKa = 4.39DD349 pKa = 3.86AVNSMLSFDD358 pKa = 3.29SRR360 pKa = 11.84EE361 pKa = 3.53IFRR364 pKa = 11.84RR365 pKa = 11.84LWADD369 pKa = 3.6CPYY372 pKa = 11.21EE373 pKa = 4.15MYY375 pKa = 8.71WQKK378 pKa = 11.07GGYY381 pKa = 8.69SFPVSLGCSKK391 pKa = 10.51RR392 pKa = 11.84ASGSGDD398 pKa = 2.8SWSGFVVIEE407 pKa = 3.93EE408 pKa = 4.52SPVTFSMCVEE418 pKa = 3.64QKK420 pKa = 10.49YY421 pKa = 9.03RR422 pKa = 11.84QDD424 pKa = 3.23QDD426 pKa = 4.28DD427 pKa = 4.76EE428 pKa = 5.37YY429 pKa = 11.78SVDD432 pKa = 3.49TSKK435 pKa = 9.75TGAACSAALFARR447 pKa = 11.84SIVNDD452 pKa = 3.72ADD454 pKa = 3.54NSGLSDD460 pKa = 3.76YY461 pKa = 11.43DD462 pKa = 3.3RR463 pKa = 11.84LVYY466 pKa = 10.79YY467 pKa = 10.41KK468 pKa = 10.72DD469 pKa = 4.03RR470 pKa = 11.84ICNEE474 pKa = 3.43VVYY477 pKa = 10.99NEE479 pKa = 4.2EE480 pKa = 3.76ARR482 pKa = 11.84QNSEE486 pKa = 4.26TYY488 pKa = 9.87PDD490 pKa = 3.71RR491 pKa = 11.84GPWALIYY498 pKa = 11.08VFDD501 pKa = 4.52QNPDD505 pKa = 3.43TNVVCEE511 pKa = 4.79GYY513 pKa = 10.22SEE515 pKa = 4.36AFQYY519 pKa = 11.24LCGLTEE525 pKa = 4.02FQSRR529 pKa = 11.84KK530 pKa = 7.39IQVYY534 pKa = 9.47SVSGMFGGGTGAGPHH549 pKa = 6.23KK550 pKa = 10.27WNIVHH555 pKa = 6.79MDD557 pKa = 3.54DD558 pKa = 4.39GLNYY562 pKa = 10.45LADD565 pKa = 3.64VTNSDD570 pKa = 4.15EE571 pKa = 4.66GTVGWDD577 pKa = 2.89GRR579 pKa = 11.84LFLKK583 pKa = 10.79GIEE586 pKa = 4.1GNVYY590 pKa = 10.49DD591 pKa = 5.06GYY593 pKa = 9.89VLSWEE598 pKa = 4.47EE599 pKa = 3.7YY600 pKa = 7.98QEE602 pKa = 4.13EE603 pKa = 4.49VEE605 pKa = 4.91EE606 pKa = 4.09EE607 pKa = 4.18DD608 pKa = 3.04GTYY611 pKa = 10.79VYY613 pKa = 9.21TYY615 pKa = 9.64PAGSVDD621 pKa = 3.48YY622 pKa = 10.67AYY624 pKa = 10.53DD625 pKa = 3.37QEE627 pKa = 4.25TRR629 pKa = 11.84ALFTDD634 pKa = 3.82EE635 pKa = 5.13EE636 pKa = 4.71LTLSSGDD643 pKa = 3.87YY644 pKa = 11.04VPGSLPPGPPAPTVDD659 pKa = 3.99EE660 pKa = 4.11IVSIEE665 pKa = 4.11EE666 pKa = 4.05VYY668 pKa = 10.76LYY670 pKa = 9.75LTDD673 pKa = 4.58KK674 pKa = 11.18IEE676 pKa = 3.97MRR678 pKa = 11.84FCAKK682 pKa = 10.37VEE684 pKa = 4.21GNIEE688 pKa = 3.76QDD690 pKa = 3.29DD691 pKa = 5.01YY692 pKa = 11.58ITFICAGRR700 pKa = 11.84TVTQTVSEE708 pKa = 4.68AEE710 pKa = 4.01TDD712 pKa = 3.57SEE714 pKa = 4.23GRR716 pKa = 11.84LVFSLEE722 pKa = 3.52LAARR726 pKa = 11.84QMTDD730 pKa = 2.32QVTYY734 pKa = 10.76FMTVDD739 pKa = 3.94GTAGTSGQYY748 pKa = 9.3SVRR751 pKa = 11.84SYY753 pKa = 11.94ADD755 pKa = 3.32TILNQSGMRR764 pKa = 11.84DD765 pKa = 3.45YY766 pKa = 11.33KK767 pKa = 11.08GLLRR771 pKa = 11.84AMLNYY776 pKa = 10.39GSYY779 pKa = 7.79TQLYY783 pKa = 8.47AGYY786 pKa = 8.05RR787 pKa = 11.84TDD789 pKa = 4.57FLAAEE794 pKa = 4.35GLYY797 pKa = 10.02TDD799 pKa = 3.92EE800 pKa = 4.73TDD802 pKa = 3.38PVLNMEE808 pKa = 4.96GPDD811 pKa = 3.67LTDD814 pKa = 3.85FAYY817 pKa = 9.42TYY819 pKa = 10.97KK820 pKa = 10.83LNSKK824 pKa = 9.1TDD826 pKa = 3.01GLTIGKK832 pKa = 10.1ASLLLGTDD840 pKa = 3.91LSLRR844 pKa = 11.84LYY846 pKa = 10.36YY847 pKa = 10.47EE848 pKa = 4.32PGEE851 pKa = 4.35GKK853 pKa = 8.54TDD855 pKa = 3.11KK856 pKa = 10.95SYY858 pKa = 11.4SIVLADD864 pKa = 4.08RR865 pKa = 11.84SGLEE869 pKa = 3.93PVLGYY874 pKa = 10.96DD875 pKa = 3.89DD876 pKa = 4.9TIGMYY881 pKa = 10.33YY882 pKa = 10.81ADD884 pKa = 4.88IEE886 pKa = 5.19HH887 pKa = 6.71ITPTQIGDD895 pKa = 3.47MFKK898 pKa = 11.01LDD900 pKa = 4.94FYY902 pKa = 11.82AEE904 pKa = 4.6DD905 pKa = 4.0GQAADD910 pKa = 4.07TPAASVTFGPLSYY923 pKa = 10.64CKK925 pKa = 10.32GVLEE929 pKa = 4.95SGSSSDD935 pKa = 4.07EE936 pKa = 3.95LVNLCRR942 pKa = 11.84ALYY945 pKa = 9.31EE946 pKa = 3.9YY947 pKa = 9.5WQIAIRR953 pKa = 11.84PLTT956 pKa = 3.64
Molecular weight: 105.2 kDa
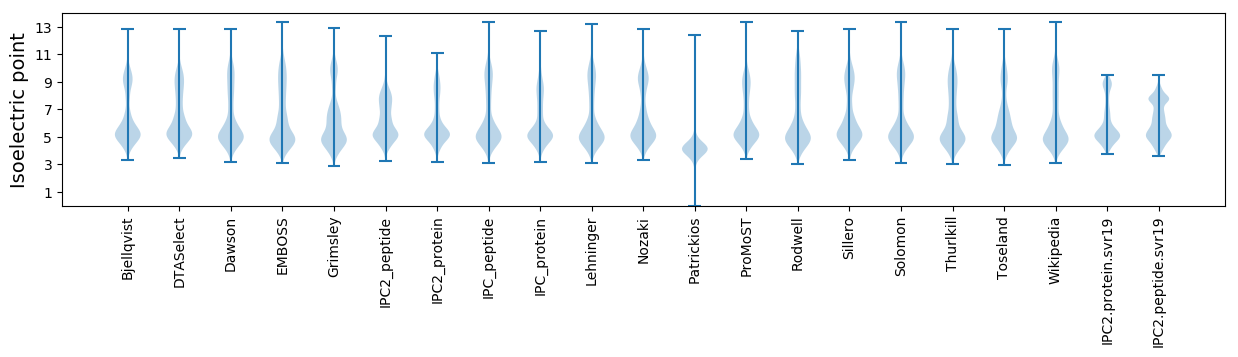
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G9K7T8|A0A1G9K7T8_9CLOT Prokaryotic molybdopterin-containing oxidoreductase family iron-sulfur binding subunit OS=Sarcina sp. DSM 11001 OX=1798184 GN=SAMN04487833_1205 PE=4 SV=1
MM1 pKa = 7.7AKK3 pKa = 8.05MTFQPKK9 pKa = 9.04KK10 pKa = 7.6RR11 pKa = 11.84QRR13 pKa = 11.84AKK15 pKa = 9.8VHH17 pKa = 5.81GFLSRR22 pKa = 11.84MSTKK26 pKa = 10.3GGRR29 pKa = 11.84KK30 pKa = 8.75VLSARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.45GRR40 pKa = 11.84KK41 pKa = 8.8QIAVV45 pKa = 3.36
MM1 pKa = 7.7AKK3 pKa = 8.05MTFQPKK9 pKa = 9.04KK10 pKa = 7.6RR11 pKa = 11.84QRR13 pKa = 11.84AKK15 pKa = 9.8VHH17 pKa = 5.81GFLSRR22 pKa = 11.84MSTKK26 pKa = 10.3GGRR29 pKa = 11.84KK30 pKa = 8.75VLSARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.45GRR40 pKa = 11.84KK41 pKa = 8.8QIAVV45 pKa = 3.36
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1153430 |
35 |
3547 |
361.9 |
40.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.266 ± 0.054 | 1.474 ± 0.018 |
6.181 ± 0.038 | 7.652 ± 0.052 |
3.942 ± 0.031 | 7.638 ± 0.049 |
1.743 ± 0.016 | 6.482 ± 0.039 |
5.655 ± 0.044 | 8.555 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.779 ± 0.025 | 3.863 ± 0.03 |
3.862 ± 0.027 | 3.079 ± 0.023 |
5.521 ± 0.047 | 6.265 ± 0.038 |
5.743 ± 0.047 | 6.499 ± 0.036 |
0.954 ± 0.015 | 3.848 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
