
Rodent stool-associated circular genome virus
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
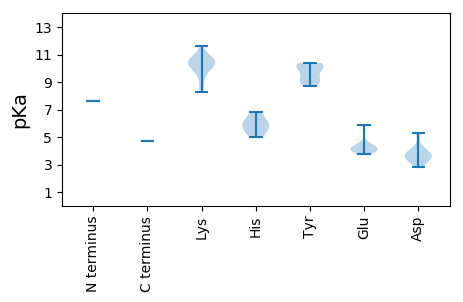
>tr|G1C9H6|G1C9H6_9VIRU ATP-dependent helicase Rep OS=Rodent stool-associated circular genome virus OX=1074214 PE=3 SV=1
MM1 pKa = 7.64CVRR4 pKa = 11.84IKK6 pKa = 8.29MTQATWWLGTKK17 pKa = 9.83FVHH20 pKa = 6.32EE21 pKa = 4.14AHH23 pKa = 6.84DD24 pKa = 3.79TEE26 pKa = 4.15EE27 pKa = 3.96WMRR30 pKa = 11.84DD31 pKa = 3.11LAEE34 pKa = 4.24SGKK37 pKa = 10.62VKK39 pKa = 10.7FITGQRR45 pKa = 11.84EE46 pKa = 4.14LCPEE50 pKa = 4.37TMRR53 pKa = 11.84HH54 pKa = 5.02HH55 pKa = 5.8VQFVVNLHH63 pKa = 6.06RR64 pKa = 11.84SQRR67 pKa = 11.84LSYY70 pKa = 10.2LKK72 pKa = 10.62KK73 pKa = 9.93IDD75 pKa = 5.13GEE77 pKa = 4.38AHH79 pKa = 5.71WEE81 pKa = 4.37SVRR84 pKa = 11.84GTPKK88 pKa = 10.09QARR91 pKa = 11.84DD92 pKa = 3.51YY93 pKa = 8.99CTKK96 pKa = 10.45EE97 pKa = 3.74DD98 pKa = 3.69TRR100 pKa = 11.84FDD102 pKa = 4.11GPWEE106 pKa = 4.03FGKK109 pKa = 10.72CLEE112 pKa = 4.49PGQRR116 pKa = 11.84RR117 pKa = 11.84GYY119 pKa = 10.3DD120 pKa = 3.16EE121 pKa = 5.59AVAAVCSGRR130 pKa = 11.84CLAEE134 pKa = 3.96VASEE138 pKa = 4.26YY139 pKa = 9.17PQVWVKK145 pKa = 10.29YY146 pKa = 10.19GRR148 pKa = 11.84GLVDD152 pKa = 3.63LRR154 pKa = 11.84KK155 pKa = 9.42QLRR158 pKa = 11.84LDD160 pKa = 3.15SDD162 pKa = 3.48RR163 pKa = 11.84RR164 pKa = 11.84QFDD167 pKa = 3.93DD168 pKa = 4.38NGPEE172 pKa = 4.04LWIFWGPSGTGKK184 pKa = 10.15SKK186 pKa = 10.58RR187 pKa = 11.84ANEE190 pKa = 4.23TWPDD194 pKa = 3.98AYY196 pKa = 9.82WKK198 pKa = 10.6IPGEE202 pKa = 4.2KK203 pKa = 8.69WWDD206 pKa = 3.48GYY208 pKa = 9.06TNQDD212 pKa = 3.14TVILDD217 pKa = 3.66DD218 pKa = 4.17FKK220 pKa = 11.6GSFMRR225 pKa = 11.84LTDD228 pKa = 3.77FQRR231 pKa = 11.84LIDD234 pKa = 5.33RR235 pKa = 11.84YY236 pKa = 8.75PLWVEE241 pKa = 4.05VKK243 pKa = 10.43GGSIPMLATRR253 pKa = 11.84YY254 pKa = 10.2VITSNEE260 pKa = 3.91APEE263 pKa = 4.02AWYY266 pKa = 10.39ASADD270 pKa = 3.35PHH272 pKa = 5.4GTVTRR277 pKa = 11.84RR278 pKa = 11.84CNDD281 pKa = 2.82FTDD284 pKa = 3.69GGRR287 pKa = 11.84RR288 pKa = 11.84IIYY291 pKa = 9.55IGPEE295 pKa = 3.89RR296 pKa = 11.84QEE298 pKa = 4.64LPDD301 pKa = 3.73FEE303 pKa = 5.84SWVGWGEE310 pKa = 3.85FARR313 pKa = 4.7
MM1 pKa = 7.64CVRR4 pKa = 11.84IKK6 pKa = 8.29MTQATWWLGTKK17 pKa = 9.83FVHH20 pKa = 6.32EE21 pKa = 4.14AHH23 pKa = 6.84DD24 pKa = 3.79TEE26 pKa = 4.15EE27 pKa = 3.96WMRR30 pKa = 11.84DD31 pKa = 3.11LAEE34 pKa = 4.24SGKK37 pKa = 10.62VKK39 pKa = 10.7FITGQRR45 pKa = 11.84EE46 pKa = 4.14LCPEE50 pKa = 4.37TMRR53 pKa = 11.84HH54 pKa = 5.02HH55 pKa = 5.8VQFVVNLHH63 pKa = 6.06RR64 pKa = 11.84SQRR67 pKa = 11.84LSYY70 pKa = 10.2LKK72 pKa = 10.62KK73 pKa = 9.93IDD75 pKa = 5.13GEE77 pKa = 4.38AHH79 pKa = 5.71WEE81 pKa = 4.37SVRR84 pKa = 11.84GTPKK88 pKa = 10.09QARR91 pKa = 11.84DD92 pKa = 3.51YY93 pKa = 8.99CTKK96 pKa = 10.45EE97 pKa = 3.74DD98 pKa = 3.69TRR100 pKa = 11.84FDD102 pKa = 4.11GPWEE106 pKa = 4.03FGKK109 pKa = 10.72CLEE112 pKa = 4.49PGQRR116 pKa = 11.84RR117 pKa = 11.84GYY119 pKa = 10.3DD120 pKa = 3.16EE121 pKa = 5.59AVAAVCSGRR130 pKa = 11.84CLAEE134 pKa = 3.96VASEE138 pKa = 4.26YY139 pKa = 9.17PQVWVKK145 pKa = 10.29YY146 pKa = 10.19GRR148 pKa = 11.84GLVDD152 pKa = 3.63LRR154 pKa = 11.84KK155 pKa = 9.42QLRR158 pKa = 11.84LDD160 pKa = 3.15SDD162 pKa = 3.48RR163 pKa = 11.84RR164 pKa = 11.84QFDD167 pKa = 3.93DD168 pKa = 4.38NGPEE172 pKa = 4.04LWIFWGPSGTGKK184 pKa = 10.15SKK186 pKa = 10.58RR187 pKa = 11.84ANEE190 pKa = 4.23TWPDD194 pKa = 3.98AYY196 pKa = 9.82WKK198 pKa = 10.6IPGEE202 pKa = 4.2KK203 pKa = 8.69WWDD206 pKa = 3.48GYY208 pKa = 9.06TNQDD212 pKa = 3.14TVILDD217 pKa = 3.66DD218 pKa = 4.17FKK220 pKa = 11.6GSFMRR225 pKa = 11.84LTDD228 pKa = 3.77FQRR231 pKa = 11.84LIDD234 pKa = 5.33RR235 pKa = 11.84YY236 pKa = 8.75PLWVEE241 pKa = 4.05VKK243 pKa = 10.43GGSIPMLATRR253 pKa = 11.84YY254 pKa = 10.2VITSNEE260 pKa = 3.91APEE263 pKa = 4.02AWYY266 pKa = 10.39ASADD270 pKa = 3.35PHH272 pKa = 5.4GTVTRR277 pKa = 11.84RR278 pKa = 11.84CNDD281 pKa = 2.82FTDD284 pKa = 3.69GGRR287 pKa = 11.84RR288 pKa = 11.84IIYY291 pKa = 9.55IGPEE295 pKa = 3.89RR296 pKa = 11.84QEE298 pKa = 4.64LPDD301 pKa = 3.73FEE303 pKa = 5.84SWVGWGEE310 pKa = 3.85FARR313 pKa = 4.7
Molecular weight: 36.5 kDa
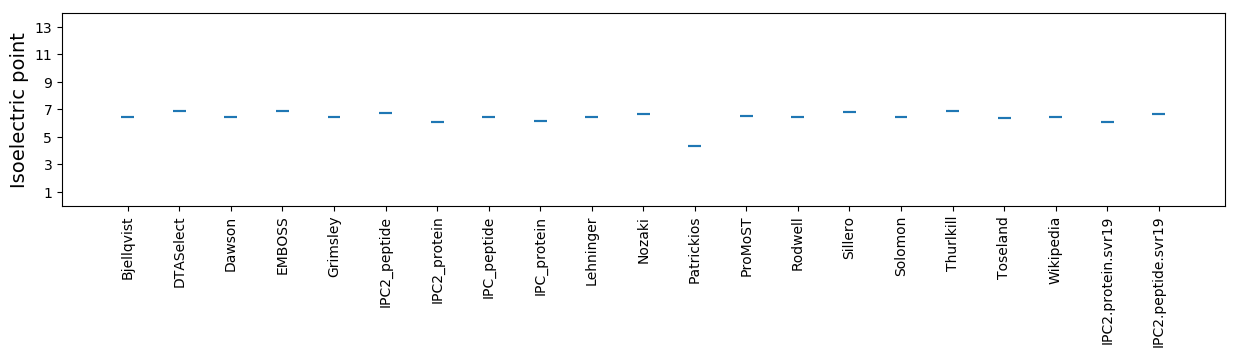
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G1C9H6|G1C9H6_9VIRU ATP-dependent helicase Rep OS=Rodent stool-associated circular genome virus OX=1074214 PE=3 SV=1
MM1 pKa = 7.64CVRR4 pKa = 11.84IKK6 pKa = 8.29MTQATWWLGTKK17 pKa = 9.83FVHH20 pKa = 6.32EE21 pKa = 4.14AHH23 pKa = 6.84DD24 pKa = 3.79TEE26 pKa = 4.15EE27 pKa = 3.96WMRR30 pKa = 11.84DD31 pKa = 3.11LAEE34 pKa = 4.24SGKK37 pKa = 10.62VKK39 pKa = 10.7FITGQRR45 pKa = 11.84EE46 pKa = 4.14LCPEE50 pKa = 4.37TMRR53 pKa = 11.84HH54 pKa = 5.02HH55 pKa = 5.8VQFVVNLHH63 pKa = 6.06RR64 pKa = 11.84SQRR67 pKa = 11.84LSYY70 pKa = 10.2LKK72 pKa = 10.62KK73 pKa = 9.93IDD75 pKa = 5.13GEE77 pKa = 4.38AHH79 pKa = 5.71WEE81 pKa = 4.37SVRR84 pKa = 11.84GTPKK88 pKa = 10.09QARR91 pKa = 11.84DD92 pKa = 3.51YY93 pKa = 8.99CTKK96 pKa = 10.45EE97 pKa = 3.74DD98 pKa = 3.69TRR100 pKa = 11.84FDD102 pKa = 4.11GPWEE106 pKa = 4.03FGKK109 pKa = 10.72CLEE112 pKa = 4.49PGQRR116 pKa = 11.84RR117 pKa = 11.84GYY119 pKa = 10.3DD120 pKa = 3.16EE121 pKa = 5.59AVAAVCSGRR130 pKa = 11.84CLAEE134 pKa = 3.96VASEE138 pKa = 4.26YY139 pKa = 9.17PQVWVKK145 pKa = 10.29YY146 pKa = 10.19GRR148 pKa = 11.84GLVDD152 pKa = 3.63LRR154 pKa = 11.84KK155 pKa = 9.42QLRR158 pKa = 11.84LDD160 pKa = 3.15SDD162 pKa = 3.48RR163 pKa = 11.84RR164 pKa = 11.84QFDD167 pKa = 3.93DD168 pKa = 4.38NGPEE172 pKa = 4.04LWIFWGPSGTGKK184 pKa = 10.15SKK186 pKa = 10.58RR187 pKa = 11.84ANEE190 pKa = 4.23TWPDD194 pKa = 3.98AYY196 pKa = 9.82WKK198 pKa = 10.6IPGEE202 pKa = 4.2KK203 pKa = 8.69WWDD206 pKa = 3.48GYY208 pKa = 9.06TNQDD212 pKa = 3.14TVILDD217 pKa = 3.66DD218 pKa = 4.17FKK220 pKa = 11.6GSFMRR225 pKa = 11.84LTDD228 pKa = 3.77FQRR231 pKa = 11.84LIDD234 pKa = 5.33RR235 pKa = 11.84YY236 pKa = 8.75PLWVEE241 pKa = 4.05VKK243 pKa = 10.43GGSIPMLATRR253 pKa = 11.84YY254 pKa = 10.2VITSNEE260 pKa = 3.91APEE263 pKa = 4.02AWYY266 pKa = 10.39ASADD270 pKa = 3.35PHH272 pKa = 5.4GTVTRR277 pKa = 11.84RR278 pKa = 11.84CNDD281 pKa = 2.82FTDD284 pKa = 3.69GGRR287 pKa = 11.84RR288 pKa = 11.84IIYY291 pKa = 9.55IGPEE295 pKa = 3.89RR296 pKa = 11.84QEE298 pKa = 4.64LPDD301 pKa = 3.73FEE303 pKa = 5.84SWVGWGEE310 pKa = 3.85FARR313 pKa = 4.7
MM1 pKa = 7.64CVRR4 pKa = 11.84IKK6 pKa = 8.29MTQATWWLGTKK17 pKa = 9.83FVHH20 pKa = 6.32EE21 pKa = 4.14AHH23 pKa = 6.84DD24 pKa = 3.79TEE26 pKa = 4.15EE27 pKa = 3.96WMRR30 pKa = 11.84DD31 pKa = 3.11LAEE34 pKa = 4.24SGKK37 pKa = 10.62VKK39 pKa = 10.7FITGQRR45 pKa = 11.84EE46 pKa = 4.14LCPEE50 pKa = 4.37TMRR53 pKa = 11.84HH54 pKa = 5.02HH55 pKa = 5.8VQFVVNLHH63 pKa = 6.06RR64 pKa = 11.84SQRR67 pKa = 11.84LSYY70 pKa = 10.2LKK72 pKa = 10.62KK73 pKa = 9.93IDD75 pKa = 5.13GEE77 pKa = 4.38AHH79 pKa = 5.71WEE81 pKa = 4.37SVRR84 pKa = 11.84GTPKK88 pKa = 10.09QARR91 pKa = 11.84DD92 pKa = 3.51YY93 pKa = 8.99CTKK96 pKa = 10.45EE97 pKa = 3.74DD98 pKa = 3.69TRR100 pKa = 11.84FDD102 pKa = 4.11GPWEE106 pKa = 4.03FGKK109 pKa = 10.72CLEE112 pKa = 4.49PGQRR116 pKa = 11.84RR117 pKa = 11.84GYY119 pKa = 10.3DD120 pKa = 3.16EE121 pKa = 5.59AVAAVCSGRR130 pKa = 11.84CLAEE134 pKa = 3.96VASEE138 pKa = 4.26YY139 pKa = 9.17PQVWVKK145 pKa = 10.29YY146 pKa = 10.19GRR148 pKa = 11.84GLVDD152 pKa = 3.63LRR154 pKa = 11.84KK155 pKa = 9.42QLRR158 pKa = 11.84LDD160 pKa = 3.15SDD162 pKa = 3.48RR163 pKa = 11.84RR164 pKa = 11.84QFDD167 pKa = 3.93DD168 pKa = 4.38NGPEE172 pKa = 4.04LWIFWGPSGTGKK184 pKa = 10.15SKK186 pKa = 10.58RR187 pKa = 11.84ANEE190 pKa = 4.23TWPDD194 pKa = 3.98AYY196 pKa = 9.82WKK198 pKa = 10.6IPGEE202 pKa = 4.2KK203 pKa = 8.69WWDD206 pKa = 3.48GYY208 pKa = 9.06TNQDD212 pKa = 3.14TVILDD217 pKa = 3.66DD218 pKa = 4.17FKK220 pKa = 11.6GSFMRR225 pKa = 11.84LTDD228 pKa = 3.77FQRR231 pKa = 11.84LIDD234 pKa = 5.33RR235 pKa = 11.84YY236 pKa = 8.75PLWVEE241 pKa = 4.05VKK243 pKa = 10.43GGSIPMLATRR253 pKa = 11.84YY254 pKa = 10.2VITSNEE260 pKa = 3.91APEE263 pKa = 4.02AWYY266 pKa = 10.39ASADD270 pKa = 3.35PHH272 pKa = 5.4GTVTRR277 pKa = 11.84RR278 pKa = 11.84CNDD281 pKa = 2.82FTDD284 pKa = 3.69GGRR287 pKa = 11.84RR288 pKa = 11.84IIYY291 pKa = 9.55IGPEE295 pKa = 3.89RR296 pKa = 11.84QEE298 pKa = 4.64LPDD301 pKa = 3.73FEE303 pKa = 5.84SWVGWGEE310 pKa = 3.85FARR313 pKa = 4.7
Molecular weight: 36.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
313 |
313 |
313 |
313.0 |
36.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.751 ± 0.0 | 2.236 ± 0.0 |
7.348 ± 0.0 | 7.668 ± 0.0 |
4.153 ± 0.0 | 8.626 ± 0.0 |
2.236 ± 0.0 | 3.834 ± 0.0 |
5.431 ± 0.0 | 6.07 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.917 ± 0.0 | 1.917 ± 0.0 |
4.792 ± 0.0 | 3.834 ± 0.0 |
8.946 ± 0.0 | 4.473 ± 0.0 |
6.07 ± 0.0 | 6.07 ± 0.0 |
5.112 ± 0.0 | 3.514 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
