
Mongoose feces-associated gemycircularvirus a
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus rhina1; Rhinolophus associated gemykibivirus 1
Average proteome isoelectric point is 7.46
Get precalculated fractions of proteins
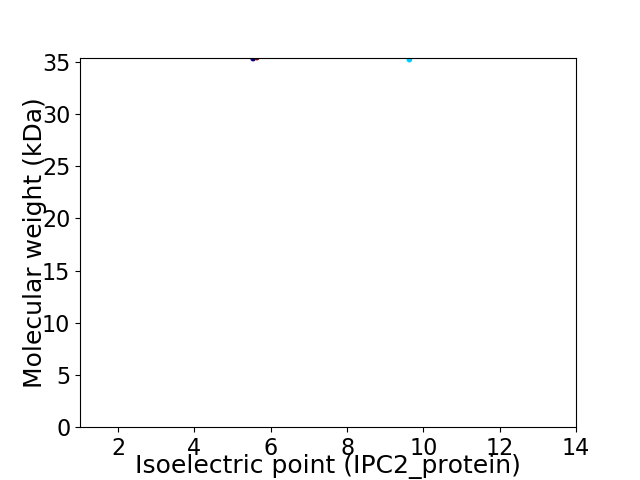
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0E3M076|A0A0E3M076_9VIRU Capsid OS=Mongoose feces-associated gemycircularvirus a OX=1634485 PE=4 SV=1
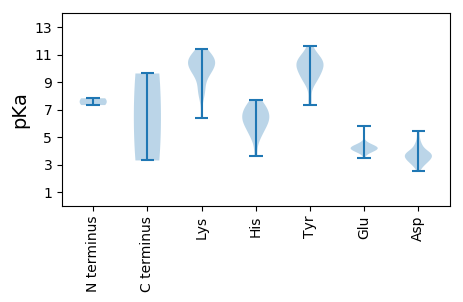
MM1 pKa = 7.32TFSFNARR8 pKa = 11.84YY9 pKa = 9.72ALLTYY14 pKa = 8.83AQCGTLDD21 pKa = 3.4PWSVVHH27 pKa = 7.01HH28 pKa = 5.91IAEE31 pKa = 4.27LRR33 pKa = 11.84GEE35 pKa = 4.3CIIARR40 pKa = 11.84EE41 pKa = 4.03AHH43 pKa = 6.55ADD45 pKa = 3.57GGTHH49 pKa = 5.97LHH51 pKa = 6.23SFIDD55 pKa = 3.87FGRR58 pKa = 11.84KK59 pKa = 7.99FRR61 pKa = 11.84SRR63 pKa = 11.84RR64 pKa = 11.84ADD66 pKa = 2.96IFDD69 pKa = 3.92VDD71 pKa = 3.78GCHH74 pKa = 6.5PNVSPTHH81 pKa = 5.44STPQAGFDD89 pKa = 3.9YY90 pKa = 10.68ACKK93 pKa = 10.71DD94 pKa = 3.18GDD96 pKa = 3.8IVAGGLSRR104 pKa = 11.84PDD106 pKa = 3.78AGPIPAKK113 pKa = 9.83HH114 pKa = 6.47DD115 pKa = 3.18RR116 pKa = 11.84WHH118 pKa = 7.68DD119 pKa = 3.08ITAAKK124 pKa = 10.16SRR126 pKa = 11.84EE127 pKa = 4.08EE128 pKa = 3.95FFQLLLEE135 pKa = 4.32HH136 pKa = 7.27APRR139 pKa = 11.84DD140 pKa = 3.47LCTSFNSLSKK150 pKa = 10.61YY151 pKa = 9.77ADD153 pKa = 2.51WMYY156 pKa = 11.4RR157 pKa = 11.84DD158 pKa = 4.69DD159 pKa = 4.36PAPYY163 pKa = 10.13KK164 pKa = 10.57HH165 pKa = 7.38DD166 pKa = 4.14DD167 pKa = 3.58QLEE170 pKa = 4.43GSLGPFPEE178 pKa = 4.39LSAWVQGSIDD188 pKa = 3.86EE189 pKa = 4.58LNSLVLYY196 pKa = 10.41GPSRR200 pKa = 11.84MGKK203 pKa = 6.35TLWARR208 pKa = 11.84SHH210 pKa = 6.62GSHH213 pKa = 7.02AYY215 pKa = 10.06FGGLFSLDD223 pKa = 3.79EE224 pKa = 4.41PLQGANYY231 pKa = 10.76AVFDD235 pKa = 5.42DD236 pKa = 4.4INGGIQFFPQYY247 pKa = 10.22KK248 pKa = 8.34WWLGHH253 pKa = 5.19QAQFYY258 pKa = 9.15ATDD261 pKa = 3.41KK262 pKa = 10.98YY263 pKa = 10.53KK264 pKa = 10.88GKK266 pKa = 10.54RR267 pKa = 11.84LIHH270 pKa = 5.94WGKK273 pKa = 8.31PAIWVANDD281 pKa = 3.79DD282 pKa = 3.54PRR284 pKa = 11.84EE285 pKa = 3.83QHH287 pKa = 5.94GADD290 pKa = 4.42RR291 pKa = 11.84EE292 pKa = 4.27WLEE295 pKa = 4.12ANCLFVYY302 pKa = 9.25VHH304 pKa = 6.6SPLYY308 pKa = 10.85SNLVV312 pKa = 3.3
MM1 pKa = 7.32TFSFNARR8 pKa = 11.84YY9 pKa = 9.72ALLTYY14 pKa = 8.83AQCGTLDD21 pKa = 3.4PWSVVHH27 pKa = 7.01HH28 pKa = 5.91IAEE31 pKa = 4.27LRR33 pKa = 11.84GEE35 pKa = 4.3CIIARR40 pKa = 11.84EE41 pKa = 4.03AHH43 pKa = 6.55ADD45 pKa = 3.57GGTHH49 pKa = 5.97LHH51 pKa = 6.23SFIDD55 pKa = 3.87FGRR58 pKa = 11.84KK59 pKa = 7.99FRR61 pKa = 11.84SRR63 pKa = 11.84RR64 pKa = 11.84ADD66 pKa = 2.96IFDD69 pKa = 3.92VDD71 pKa = 3.78GCHH74 pKa = 6.5PNVSPTHH81 pKa = 5.44STPQAGFDD89 pKa = 3.9YY90 pKa = 10.68ACKK93 pKa = 10.71DD94 pKa = 3.18GDD96 pKa = 3.8IVAGGLSRR104 pKa = 11.84PDD106 pKa = 3.78AGPIPAKK113 pKa = 9.83HH114 pKa = 6.47DD115 pKa = 3.18RR116 pKa = 11.84WHH118 pKa = 7.68DD119 pKa = 3.08ITAAKK124 pKa = 10.16SRR126 pKa = 11.84EE127 pKa = 4.08EE128 pKa = 3.95FFQLLLEE135 pKa = 4.32HH136 pKa = 7.27APRR139 pKa = 11.84DD140 pKa = 3.47LCTSFNSLSKK150 pKa = 10.61YY151 pKa = 9.77ADD153 pKa = 2.51WMYY156 pKa = 11.4RR157 pKa = 11.84DD158 pKa = 4.69DD159 pKa = 4.36PAPYY163 pKa = 10.13KK164 pKa = 10.57HH165 pKa = 7.38DD166 pKa = 4.14DD167 pKa = 3.58QLEE170 pKa = 4.43GSLGPFPEE178 pKa = 4.39LSAWVQGSIDD188 pKa = 3.86EE189 pKa = 4.58LNSLVLYY196 pKa = 10.41GPSRR200 pKa = 11.84MGKK203 pKa = 6.35TLWARR208 pKa = 11.84SHH210 pKa = 6.62GSHH213 pKa = 7.02AYY215 pKa = 10.06FGGLFSLDD223 pKa = 3.79EE224 pKa = 4.41PLQGANYY231 pKa = 10.76AVFDD235 pKa = 5.42DD236 pKa = 4.4INGGIQFFPQYY247 pKa = 10.22KK248 pKa = 8.34WWLGHH253 pKa = 5.19QAQFYY258 pKa = 9.15ATDD261 pKa = 3.41KK262 pKa = 10.98YY263 pKa = 10.53KK264 pKa = 10.88GKK266 pKa = 10.54RR267 pKa = 11.84LIHH270 pKa = 5.94WGKK273 pKa = 8.31PAIWVANDD281 pKa = 3.79DD282 pKa = 3.54PRR284 pKa = 11.84EE285 pKa = 3.83QHH287 pKa = 5.94GADD290 pKa = 4.42RR291 pKa = 11.84EE292 pKa = 4.27WLEE295 pKa = 4.12ANCLFVYY302 pKa = 9.25VHH304 pKa = 6.6SPLYY308 pKa = 10.85SNLVV312 pKa = 3.3
Molecular weight: 35.31 kDa
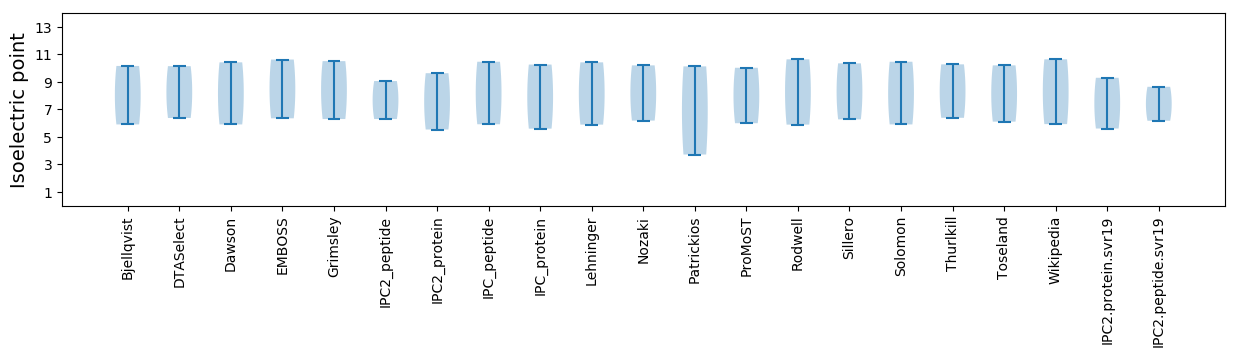
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0E3M076|A0A0E3M076_9VIRU Capsid OS=Mongoose feces-associated gemycircularvirus a OX=1634485 PE=4 SV=1
MM1 pKa = 7.82AFRR4 pKa = 11.84SRR6 pKa = 11.84YY7 pKa = 7.78SSRR10 pKa = 11.84SRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84PLRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84YY20 pKa = 9.6RR21 pKa = 11.84KK22 pKa = 8.76IRR24 pKa = 11.84RR25 pKa = 11.84SFRR28 pKa = 11.84RR29 pKa = 11.84GARR32 pKa = 11.84RR33 pKa = 11.84YY34 pKa = 7.33MRR36 pKa = 11.84RR37 pKa = 11.84SMTRR41 pKa = 11.84RR42 pKa = 11.84KK43 pKa = 9.89VADD46 pKa = 3.27IASTKK51 pKa = 10.74CKK53 pKa = 10.89DD54 pKa = 3.45NMLALPLTGDD64 pKa = 3.7LGTPGVAGEE73 pKa = 3.9AAIVQGAFISGYY85 pKa = 9.0LFCPTARR92 pKa = 11.84SSEE95 pKa = 3.92WTEE98 pKa = 3.93GEE100 pKa = 4.25RR101 pKa = 11.84EE102 pKa = 4.15VTRR105 pKa = 11.84QQSRR109 pKa = 11.84VFARR113 pKa = 11.84GYY115 pKa = 9.95KK116 pKa = 9.44EE117 pKa = 4.25RR118 pKa = 11.84INIVTSDD125 pKa = 3.38STNWIWRR132 pKa = 11.84RR133 pKa = 11.84IVFSTHH139 pKa = 3.59NRR141 pKa = 11.84LWEE144 pKa = 4.22AFPPGTVEE152 pKa = 4.27KK153 pKa = 9.93YY154 pKa = 9.28YY155 pKa = 10.87EE156 pKa = 4.57GGTGVFQPGQTRR168 pKa = 11.84AMWNFAPSLGGAPAAAVNYY187 pKa = 10.36AVFEE191 pKa = 4.34GEE193 pKa = 5.8RR194 pKa = 11.84GNDD197 pKa = 2.88WLNFMNATTNKK208 pKa = 9.93KK209 pKa = 9.63FIKK212 pKa = 10.51VMSDD216 pKa = 2.89VTQKK220 pKa = 11.4LEE222 pKa = 4.09GTNDD226 pKa = 3.23RR227 pKa = 11.84PRR229 pKa = 11.84QYY231 pKa = 10.84HH232 pKa = 5.31FNRR235 pKa = 11.84WYY237 pKa = 10.12PFNKK241 pKa = 9.67NFTYY245 pKa = 10.62NEE247 pKa = 4.16KK248 pKa = 10.5EE249 pKa = 4.08RR250 pKa = 11.84GDD252 pKa = 3.79TKK254 pKa = 10.68PSQDD258 pKa = 3.72YY259 pKa = 10.36QSKK262 pKa = 10.22FSSNEE267 pKa = 3.46IGSMGDD273 pKa = 3.4VYY275 pKa = 11.64VLDD278 pKa = 5.11LFNSANGEE286 pKa = 4.07VGNQLGFQCHH296 pKa = 4.73GTYY299 pKa = 9.9YY300 pKa = 9.42WHH302 pKa = 7.23EE303 pKa = 4.14KK304 pKa = 9.62
MM1 pKa = 7.82AFRR4 pKa = 11.84SRR6 pKa = 11.84YY7 pKa = 7.78SSRR10 pKa = 11.84SRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84PLRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84YY20 pKa = 9.6RR21 pKa = 11.84KK22 pKa = 8.76IRR24 pKa = 11.84RR25 pKa = 11.84SFRR28 pKa = 11.84RR29 pKa = 11.84GARR32 pKa = 11.84RR33 pKa = 11.84YY34 pKa = 7.33MRR36 pKa = 11.84RR37 pKa = 11.84SMTRR41 pKa = 11.84RR42 pKa = 11.84KK43 pKa = 9.89VADD46 pKa = 3.27IASTKK51 pKa = 10.74CKK53 pKa = 10.89DD54 pKa = 3.45NMLALPLTGDD64 pKa = 3.7LGTPGVAGEE73 pKa = 3.9AAIVQGAFISGYY85 pKa = 9.0LFCPTARR92 pKa = 11.84SSEE95 pKa = 3.92WTEE98 pKa = 3.93GEE100 pKa = 4.25RR101 pKa = 11.84EE102 pKa = 4.15VTRR105 pKa = 11.84QQSRR109 pKa = 11.84VFARR113 pKa = 11.84GYY115 pKa = 9.95KK116 pKa = 9.44EE117 pKa = 4.25RR118 pKa = 11.84INIVTSDD125 pKa = 3.38STNWIWRR132 pKa = 11.84RR133 pKa = 11.84IVFSTHH139 pKa = 3.59NRR141 pKa = 11.84LWEE144 pKa = 4.22AFPPGTVEE152 pKa = 4.27KK153 pKa = 9.93YY154 pKa = 9.28YY155 pKa = 10.87EE156 pKa = 4.57GGTGVFQPGQTRR168 pKa = 11.84AMWNFAPSLGGAPAAAVNYY187 pKa = 10.36AVFEE191 pKa = 4.34GEE193 pKa = 5.8RR194 pKa = 11.84GNDD197 pKa = 2.88WLNFMNATTNKK208 pKa = 9.93KK209 pKa = 9.63FIKK212 pKa = 10.51VMSDD216 pKa = 2.89VTQKK220 pKa = 11.4LEE222 pKa = 4.09GTNDD226 pKa = 3.23RR227 pKa = 11.84PRR229 pKa = 11.84QYY231 pKa = 10.84HH232 pKa = 5.31FNRR235 pKa = 11.84WYY237 pKa = 10.12PFNKK241 pKa = 9.67NFTYY245 pKa = 10.62NEE247 pKa = 4.16KK248 pKa = 10.5EE249 pKa = 4.08RR250 pKa = 11.84GDD252 pKa = 3.79TKK254 pKa = 10.68PSQDD258 pKa = 3.72YY259 pKa = 10.36QSKK262 pKa = 10.22FSSNEE267 pKa = 3.46IGSMGDD273 pKa = 3.4VYY275 pKa = 11.64VLDD278 pKa = 5.11LFNSANGEE286 pKa = 4.07VGNQLGFQCHH296 pKa = 4.73GTYY299 pKa = 9.9YY300 pKa = 9.42WHH302 pKa = 7.23EE303 pKa = 4.14KK304 pKa = 9.62
Molecular weight: 35.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
616 |
304 |
312 |
308.0 |
35.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.117 ± 0.815 | 1.461 ± 0.32 |
6.006 ± 1.611 | 4.87 ± 0.487 |
5.844 ± 0.052 | 8.442 ± 0.075 |
3.409 ± 1.412 | 3.734 ± 0.3 |
4.383 ± 0.372 | 6.494 ± 1.496 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.786 ± 0.571 | 4.545 ± 1.15 |
4.87 ± 0.622 | 3.571 ± 0.032 |
8.442 ± 2.072 | 6.981 ± 0.173 |
4.87 ± 1.153 | 4.545 ± 0.484 |
2.922 ± 0.196 | 4.708 ± 0.153 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
