
Azoarcus indigens
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Rhodocyclales; Zoogloeaceae; Azoarcus
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
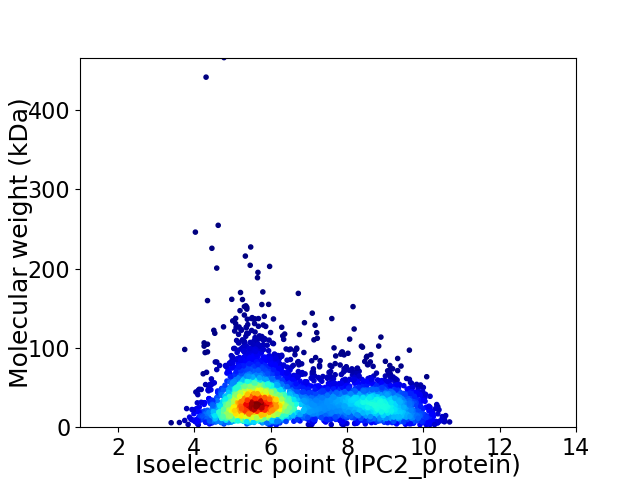
Virtual 2D-PAGE plot for 4840 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R6DXG2|A0A4R6DXG2_9RHOO PqqA peptide cyclase OS=Azoarcus indigens OX=29545 GN=pqqE PE=3 SV=1
MM1 pKa = 7.69SEE3 pKa = 4.52VIDD6 pKa = 3.84TPEE9 pKa = 3.97ILVFTDD15 pKa = 3.54SAANKK20 pKa = 9.08VRR22 pKa = 11.84EE23 pKa = 4.59LIDD26 pKa = 3.62EE27 pKa = 4.54EE28 pKa = 4.86GNPEE32 pKa = 3.75LKK34 pKa = 10.69LRR36 pKa = 11.84VFVTGGGCSGFQYY49 pKa = 10.93GFTFDD54 pKa = 4.28EE55 pKa = 4.93EE56 pKa = 4.59VNEE59 pKa = 5.25DD60 pKa = 3.38DD61 pKa = 4.2TTFEE65 pKa = 4.48KK66 pKa = 10.86NGVMLLIDD74 pKa = 3.94PMSYY78 pKa = 10.31QYY80 pKa = 11.58LVGAEE85 pKa = 3.74IDD87 pKa = 3.65YY88 pKa = 11.47SEE90 pKa = 4.55GLEE93 pKa = 4.01GSQFVIRR100 pKa = 11.84NPNATSTCGCGSSFSAA116 pKa = 4.79
MM1 pKa = 7.69SEE3 pKa = 4.52VIDD6 pKa = 3.84TPEE9 pKa = 3.97ILVFTDD15 pKa = 3.54SAANKK20 pKa = 9.08VRR22 pKa = 11.84EE23 pKa = 4.59LIDD26 pKa = 3.62EE27 pKa = 4.54EE28 pKa = 4.86GNPEE32 pKa = 3.75LKK34 pKa = 10.69LRR36 pKa = 11.84VFVTGGGCSGFQYY49 pKa = 10.93GFTFDD54 pKa = 4.28EE55 pKa = 4.93EE56 pKa = 4.59VNEE59 pKa = 5.25DD60 pKa = 3.38DD61 pKa = 4.2TTFEE65 pKa = 4.48KK66 pKa = 10.86NGVMLLIDD74 pKa = 3.94PMSYY78 pKa = 10.31QYY80 pKa = 11.58LVGAEE85 pKa = 3.74IDD87 pKa = 3.65YY88 pKa = 11.47SEE90 pKa = 4.55GLEE93 pKa = 4.01GSQFVIRR100 pKa = 11.84NPNATSTCGCGSSFSAA116 pKa = 4.79
Molecular weight: 12.66 kDa
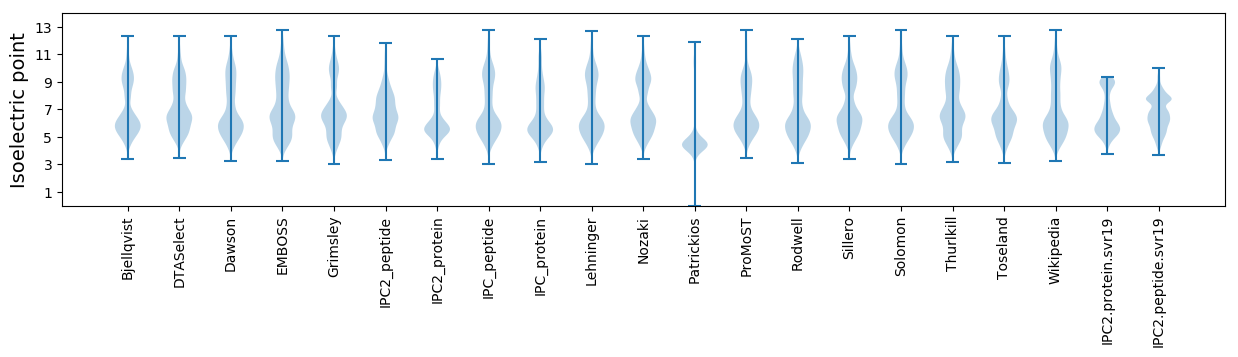
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R6E772|A0A4R6E772_9RHOO Anthranilate synthase component II OS=Azoarcus indigens OX=29545 GN=C7389_104127 PE=4 SV=1
MM1 pKa = 7.28KK2 pKa = 10.0AAAGGVMKK10 pKa = 10.47AYY12 pKa = 9.8RR13 pKa = 11.84RR14 pKa = 11.84CVFVALALTLIVVVAGAYY32 pKa = 9.69VRR34 pKa = 11.84LSDD37 pKa = 5.29AGLGCPDD44 pKa = 3.01WPGCYY49 pKa = 9.67GQLSPHH55 pKa = 6.57HH56 pKa = 6.52AADD59 pKa = 5.85DD60 pKa = 3.54ILAAHH65 pKa = 7.02AADD68 pKa = 4.29PYY70 pKa = 11.34GPVSLAKK77 pKa = 9.62AWKK80 pKa = 9.69EE81 pKa = 3.69MIHH84 pKa = 6.81RR85 pKa = 11.84YY86 pKa = 8.86LAGGLGLLLIAIAVLAWRR104 pKa = 11.84LRR106 pKa = 11.84GEE108 pKa = 4.19RR109 pKa = 11.84EE110 pKa = 3.71GAARR114 pKa = 11.84SAPVAALLLLAVVVLQGLLGKK135 pKa = 8.19WTVTLLLKK143 pKa = 10.13PAIVTAHH150 pKa = 7.09LLGGMAVLGLLAWIALRR167 pKa = 11.84ASGVRR172 pKa = 11.84RR173 pKa = 11.84VAAAPATVAALRR185 pKa = 11.84LGLLLLVGQIALGGWTSTNYY205 pKa = 10.47AALACPDD212 pKa = 3.98FPTCHH217 pKa = 6.31GSLWPEE223 pKa = 3.22TDD225 pKa = 3.07YY226 pKa = 11.74RR227 pKa = 11.84NAFHH231 pKa = 6.78VVRR234 pKa = 11.84EE235 pKa = 4.26LGMTADD241 pKa = 4.46GEE243 pKa = 4.44PLSFAALTAIHH254 pKa = 6.17WMHH257 pKa = 7.34RR258 pKa = 11.84IGAALVGGFLLVVGWRR274 pKa = 11.84LSRR277 pKa = 11.84QSSSALMGAALIGGVLLQVTIGIANVLFSLPLGLAVAHH315 pKa = 6.17NAGAALLLGILVWINHH331 pKa = 5.53GFCLRR336 pKa = 11.84TAPAGLRR343 pKa = 11.84FRR345 pKa = 11.84ARR347 pKa = 11.84PGLVRR352 pKa = 11.84IGWSAAAHH360 pKa = 5.22SHH362 pKa = 5.43GQVLPFRR369 pKa = 11.84PTAMRR374 pKa = 11.84KK375 pKa = 9.55RR376 pKa = 11.84GHH378 pKa = 6.08AA379 pKa = 3.46
MM1 pKa = 7.28KK2 pKa = 10.0AAAGGVMKK10 pKa = 10.47AYY12 pKa = 9.8RR13 pKa = 11.84RR14 pKa = 11.84CVFVALALTLIVVVAGAYY32 pKa = 9.69VRR34 pKa = 11.84LSDD37 pKa = 5.29AGLGCPDD44 pKa = 3.01WPGCYY49 pKa = 9.67GQLSPHH55 pKa = 6.57HH56 pKa = 6.52AADD59 pKa = 5.85DD60 pKa = 3.54ILAAHH65 pKa = 7.02AADD68 pKa = 4.29PYY70 pKa = 11.34GPVSLAKK77 pKa = 9.62AWKK80 pKa = 9.69EE81 pKa = 3.69MIHH84 pKa = 6.81RR85 pKa = 11.84YY86 pKa = 8.86LAGGLGLLLIAIAVLAWRR104 pKa = 11.84LRR106 pKa = 11.84GEE108 pKa = 4.19RR109 pKa = 11.84EE110 pKa = 3.71GAARR114 pKa = 11.84SAPVAALLLLAVVVLQGLLGKK135 pKa = 8.19WTVTLLLKK143 pKa = 10.13PAIVTAHH150 pKa = 7.09LLGGMAVLGLLAWIALRR167 pKa = 11.84ASGVRR172 pKa = 11.84RR173 pKa = 11.84VAAAPATVAALRR185 pKa = 11.84LGLLLLVGQIALGGWTSTNYY205 pKa = 10.47AALACPDD212 pKa = 3.98FPTCHH217 pKa = 6.31GSLWPEE223 pKa = 3.22TDD225 pKa = 3.07YY226 pKa = 11.74RR227 pKa = 11.84NAFHH231 pKa = 6.78VVRR234 pKa = 11.84EE235 pKa = 4.26LGMTADD241 pKa = 4.46GEE243 pKa = 4.44PLSFAALTAIHH254 pKa = 6.17WMHH257 pKa = 7.34RR258 pKa = 11.84IGAALVGGFLLVVGWRR274 pKa = 11.84LSRR277 pKa = 11.84QSSSALMGAALIGGVLLQVTIGIANVLFSLPLGLAVAHH315 pKa = 6.17NAGAALLLGILVWINHH331 pKa = 5.53GFCLRR336 pKa = 11.84TAPAGLRR343 pKa = 11.84FRR345 pKa = 11.84ARR347 pKa = 11.84PGLVRR352 pKa = 11.84IGWSAAAHH360 pKa = 5.22SHH362 pKa = 5.43GQVLPFRR369 pKa = 11.84PTAMRR374 pKa = 11.84KK375 pKa = 9.55RR376 pKa = 11.84GHH378 pKa = 6.08AA379 pKa = 3.46
Molecular weight: 39.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1613751 |
28 |
4533 |
333.4 |
36.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.37 ± 0.053 | 0.93 ± 0.011 |
5.22 ± 0.021 | 5.868 ± 0.039 |
3.453 ± 0.021 | 8.657 ± 0.045 |
2.173 ± 0.018 | 4.175 ± 0.026 |
2.753 ± 0.033 | 11.379 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.226 ± 0.017 | 2.475 ± 0.025 |
5.308 ± 0.03 | 3.571 ± 0.02 |
7.576 ± 0.043 | 5.192 ± 0.039 |
4.681 ± 0.037 | 7.273 ± 0.025 |
1.399 ± 0.015 | 2.32 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
