
Gammapapillomavirus 9
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
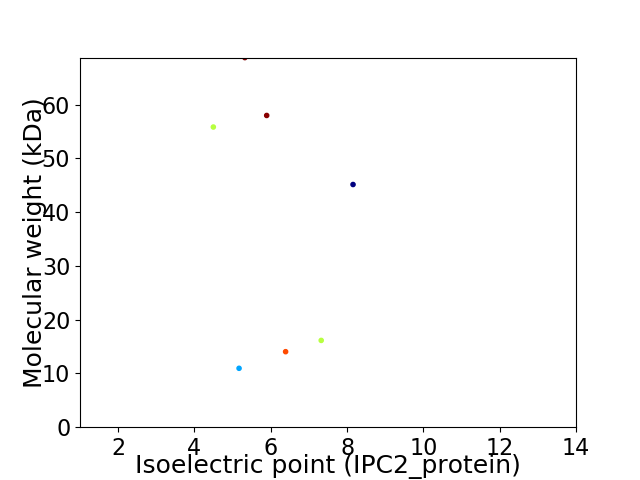
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D2ALP6|A0A2D2ALP6_9PAPI Major capsid protein L1 OS=Gammapapillomavirus 9 OX=1175851 GN=L1 PE=3 SV=1
MM1 pKa = 6.99NRR3 pKa = 11.84SRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.06RR8 pKa = 11.84ASPTEE13 pKa = 4.19LYY15 pKa = 10.22KK16 pKa = 10.83SCAMGGDD23 pKa = 4.97CIPDD27 pKa = 3.29VKK29 pKa = 11.11NKK31 pKa = 10.15IEE33 pKa = 4.16GTTLADD39 pKa = 2.99ILLKK43 pKa = 11.03VFGSVIYY50 pKa = 10.49LGNLGIGTGRR60 pKa = 11.84GSGGQYY66 pKa = 10.53GYY68 pKa = 9.21TPFGGTKK75 pKa = 8.31PTTSATPLRR84 pKa = 11.84PSIPTDD90 pKa = 3.47TVVTGDD96 pKa = 3.92ILPVTPYY103 pKa = 11.05DD104 pKa = 3.69SAIVPLTDD112 pKa = 4.25GLPEE116 pKa = 3.87PAIIDD121 pKa = 3.92TPGVGPGLPTDD132 pKa = 3.76SVTVTTTIDD141 pKa = 4.22PISEE145 pKa = 4.03VTGVGEE151 pKa = 4.39HH152 pKa = 6.97PNVIYY157 pKa = 10.94NADD160 pKa = 3.38NVAQLDD166 pKa = 4.17VQLQPPPPKK175 pKa = 10.16KK176 pKa = 10.68VILDD180 pKa = 3.63ASIHH184 pKa = 4.58NTEE187 pKa = 4.46TNIHH191 pKa = 4.7THH193 pKa = 6.3ASHH196 pKa = 6.8VDD198 pKa = 2.75SDD200 pKa = 4.09YY201 pKa = 11.48TVFVDD206 pKa = 3.89AQFHH210 pKa = 6.4GEE212 pKa = 4.04HH213 pKa = 6.65IGPVEE218 pKa = 4.21EE219 pKa = 6.17IEE221 pKa = 4.15LQSINLRR228 pKa = 11.84EE229 pKa = 3.97EE230 pKa = 4.09FEE232 pKa = 4.12IDD234 pKa = 3.22EE235 pKa = 5.19GPLKK239 pKa = 10.0STPLSSRR246 pKa = 11.84LINRR250 pKa = 11.84TRR252 pKa = 11.84DD253 pKa = 3.09LYY255 pKa = 11.18HH256 pKa = 6.95RR257 pKa = 11.84FVEE260 pKa = 4.31QVPTQKK266 pKa = 10.52EE267 pKa = 4.45VVLSRR272 pKa = 11.84PGVTYY277 pKa = 10.4EE278 pKa = 4.16FEE280 pKa = 4.5NPAFDD285 pKa = 5.1PDD287 pKa = 4.09VIDD290 pKa = 3.46VFEE293 pKa = 5.3RR294 pKa = 11.84EE295 pKa = 4.21VQEE298 pKa = 4.44VARR301 pKa = 11.84TAPLEE306 pKa = 4.16DD307 pKa = 3.66TQDD310 pKa = 3.71VIRR313 pKa = 11.84LGDD316 pKa = 3.45TRR318 pKa = 11.84FDD320 pKa = 3.42EE321 pKa = 4.72SPRR324 pKa = 11.84GTVRR328 pKa = 11.84VSRR331 pKa = 11.84LGQRR335 pKa = 11.84SGMITRR341 pKa = 11.84SGLQIGKK348 pKa = 9.33RR349 pKa = 11.84VHH351 pKa = 6.69FYY353 pKa = 11.36YY354 pKa = 10.5DD355 pKa = 3.04VSPIAGEE362 pKa = 4.26AIEE365 pKa = 4.32LRR367 pKa = 11.84TVGEE371 pKa = 4.23YY372 pKa = 10.01SHH374 pKa = 6.86EE375 pKa = 4.17SSIVDD380 pKa = 3.62EE381 pKa = 4.64LTTSSFVNPFEE392 pKa = 4.16MPIDD396 pKa = 3.9SSLQFSDD403 pKa = 5.57DD404 pKa = 3.66MLLEE408 pKa = 4.35SATEE412 pKa = 4.06DD413 pKa = 3.61FSNSHH418 pKa = 6.84VILTNTEE425 pKa = 3.81TDD427 pKa = 3.11GDD429 pKa = 4.51AIDD432 pKa = 4.78IPVNLPNEE440 pKa = 4.29GVKK443 pKa = 10.59VFVDD447 pKa = 5.67DD448 pKa = 3.57YY449 pKa = 9.16TQPLFVSHH457 pKa = 7.78PINVDD462 pKa = 3.03TSVIDD467 pKa = 3.77FDD469 pKa = 4.19TPLIPFQPSFGLDD482 pKa = 3.2VNYY485 pKa = 10.14FDD487 pKa = 5.84YY488 pKa = 11.07DD489 pKa = 3.36IHH491 pKa = 7.22PSLLLRR497 pKa = 11.84KK498 pKa = 9.26RR499 pKa = 11.84KK500 pKa = 9.87YY501 pKa = 10.18PVFFTLL507 pKa = 4.95
MM1 pKa = 6.99NRR3 pKa = 11.84SRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.06RR8 pKa = 11.84ASPTEE13 pKa = 4.19LYY15 pKa = 10.22KK16 pKa = 10.83SCAMGGDD23 pKa = 4.97CIPDD27 pKa = 3.29VKK29 pKa = 11.11NKK31 pKa = 10.15IEE33 pKa = 4.16GTTLADD39 pKa = 2.99ILLKK43 pKa = 11.03VFGSVIYY50 pKa = 10.49LGNLGIGTGRR60 pKa = 11.84GSGGQYY66 pKa = 10.53GYY68 pKa = 9.21TPFGGTKK75 pKa = 8.31PTTSATPLRR84 pKa = 11.84PSIPTDD90 pKa = 3.47TVVTGDD96 pKa = 3.92ILPVTPYY103 pKa = 11.05DD104 pKa = 3.69SAIVPLTDD112 pKa = 4.25GLPEE116 pKa = 3.87PAIIDD121 pKa = 3.92TPGVGPGLPTDD132 pKa = 3.76SVTVTTTIDD141 pKa = 4.22PISEE145 pKa = 4.03VTGVGEE151 pKa = 4.39HH152 pKa = 6.97PNVIYY157 pKa = 10.94NADD160 pKa = 3.38NVAQLDD166 pKa = 4.17VQLQPPPPKK175 pKa = 10.16KK176 pKa = 10.68VILDD180 pKa = 3.63ASIHH184 pKa = 4.58NTEE187 pKa = 4.46TNIHH191 pKa = 4.7THH193 pKa = 6.3ASHH196 pKa = 6.8VDD198 pKa = 2.75SDD200 pKa = 4.09YY201 pKa = 11.48TVFVDD206 pKa = 3.89AQFHH210 pKa = 6.4GEE212 pKa = 4.04HH213 pKa = 6.65IGPVEE218 pKa = 4.21EE219 pKa = 6.17IEE221 pKa = 4.15LQSINLRR228 pKa = 11.84EE229 pKa = 3.97EE230 pKa = 4.09FEE232 pKa = 4.12IDD234 pKa = 3.22EE235 pKa = 5.19GPLKK239 pKa = 10.0STPLSSRR246 pKa = 11.84LINRR250 pKa = 11.84TRR252 pKa = 11.84DD253 pKa = 3.09LYY255 pKa = 11.18HH256 pKa = 6.95RR257 pKa = 11.84FVEE260 pKa = 4.31QVPTQKK266 pKa = 10.52EE267 pKa = 4.45VVLSRR272 pKa = 11.84PGVTYY277 pKa = 10.4EE278 pKa = 4.16FEE280 pKa = 4.5NPAFDD285 pKa = 5.1PDD287 pKa = 4.09VIDD290 pKa = 3.46VFEE293 pKa = 5.3RR294 pKa = 11.84EE295 pKa = 4.21VQEE298 pKa = 4.44VARR301 pKa = 11.84TAPLEE306 pKa = 4.16DD307 pKa = 3.66TQDD310 pKa = 3.71VIRR313 pKa = 11.84LGDD316 pKa = 3.45TRR318 pKa = 11.84FDD320 pKa = 3.42EE321 pKa = 4.72SPRR324 pKa = 11.84GTVRR328 pKa = 11.84VSRR331 pKa = 11.84LGQRR335 pKa = 11.84SGMITRR341 pKa = 11.84SGLQIGKK348 pKa = 9.33RR349 pKa = 11.84VHH351 pKa = 6.69FYY353 pKa = 11.36YY354 pKa = 10.5DD355 pKa = 3.04VSPIAGEE362 pKa = 4.26AIEE365 pKa = 4.32LRR367 pKa = 11.84TVGEE371 pKa = 4.23YY372 pKa = 10.01SHH374 pKa = 6.86EE375 pKa = 4.17SSIVDD380 pKa = 3.62EE381 pKa = 4.64LTTSSFVNPFEE392 pKa = 4.16MPIDD396 pKa = 3.9SSLQFSDD403 pKa = 5.57DD404 pKa = 3.66MLLEE408 pKa = 4.35SATEE412 pKa = 4.06DD413 pKa = 3.61FSNSHH418 pKa = 6.84VILTNTEE425 pKa = 3.81TDD427 pKa = 3.11GDD429 pKa = 4.51AIDD432 pKa = 4.78IPVNLPNEE440 pKa = 4.29GVKK443 pKa = 10.59VFVDD447 pKa = 5.67DD448 pKa = 3.57YY449 pKa = 9.16TQPLFVSHH457 pKa = 7.78PINVDD462 pKa = 3.03TSVIDD467 pKa = 3.77FDD469 pKa = 4.19TPLIPFQPSFGLDD482 pKa = 3.2VNYY485 pKa = 10.14FDD487 pKa = 5.84YY488 pKa = 11.07DD489 pKa = 3.36IHH491 pKa = 7.22PSLLLRR497 pKa = 11.84KK498 pKa = 9.26RR499 pKa = 11.84KK500 pKa = 9.87YY501 pKa = 10.18PVFFTLL507 pKa = 4.95
Molecular weight: 55.85 kDa
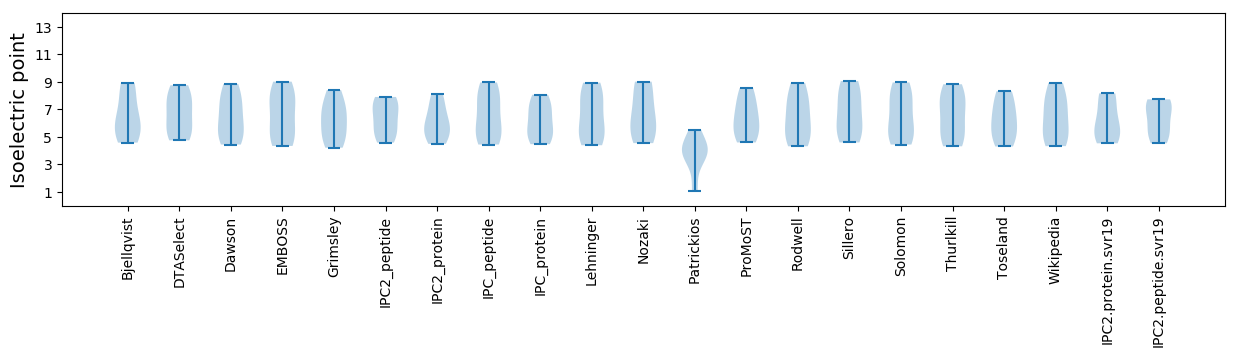
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D2ALI8|A0A2D2ALI8_9PAPI Protein E7 OS=Gammapapillomavirus 9 OX=1175851 GN=E7 PE=3 SV=1
MM1 pKa = 6.91EE2 pKa = 4.86TLRR5 pKa = 11.84PTDD8 pKa = 3.62LKK10 pKa = 11.06SYY12 pKa = 8.9CTIFGIDD19 pKa = 3.71FFEE22 pKa = 5.15LHH24 pKa = 6.27LQCIFCNCYY33 pKa = 10.42LSLVDD38 pKa = 5.81LADD41 pKa = 5.18FYY43 pKa = 10.63TKK45 pKa = 10.29CLCLVWRR52 pKa = 11.84VGKK55 pKa = 10.13AHH57 pKa = 7.09ACCAKK62 pKa = 10.16CLCLSAKK69 pKa = 10.42CEE71 pKa = 4.23LEE73 pKa = 4.04RR74 pKa = 11.84HH75 pKa = 4.99VQCSAKK81 pKa = 10.18VVNLHH86 pKa = 6.34SLTNKK91 pKa = 9.15PLCEE95 pKa = 3.94LKK97 pKa = 10.77VRR99 pKa = 11.84CCQCLALLDD108 pKa = 4.17LQEE111 pKa = 4.62KK112 pKa = 9.9CDD114 pKa = 3.71LVARR118 pKa = 11.84GKK120 pKa = 9.98DD121 pKa = 3.19AYY123 pKa = 10.58LVRR126 pKa = 11.84GNWRR130 pKa = 11.84APCRR134 pKa = 11.84KK135 pKa = 10.06CIDD138 pKa = 3.53RR139 pKa = 11.84EE140 pKa = 4.05II141 pKa = 5.35
MM1 pKa = 6.91EE2 pKa = 4.86TLRR5 pKa = 11.84PTDD8 pKa = 3.62LKK10 pKa = 11.06SYY12 pKa = 8.9CTIFGIDD19 pKa = 3.71FFEE22 pKa = 5.15LHH24 pKa = 6.27LQCIFCNCYY33 pKa = 10.42LSLVDD38 pKa = 5.81LADD41 pKa = 5.18FYY43 pKa = 10.63TKK45 pKa = 10.29CLCLVWRR52 pKa = 11.84VGKK55 pKa = 10.13AHH57 pKa = 7.09ACCAKK62 pKa = 10.16CLCLSAKK69 pKa = 10.42CEE71 pKa = 4.23LEE73 pKa = 4.04RR74 pKa = 11.84HH75 pKa = 4.99VQCSAKK81 pKa = 10.18VVNLHH86 pKa = 6.34SLTNKK91 pKa = 9.15PLCEE95 pKa = 3.94LKK97 pKa = 10.77VRR99 pKa = 11.84CCQCLALLDD108 pKa = 4.17LQEE111 pKa = 4.62KK112 pKa = 9.9CDD114 pKa = 3.71LVARR118 pKa = 11.84GKK120 pKa = 9.98DD121 pKa = 3.19AYY123 pKa = 10.58LVRR126 pKa = 11.84GNWRR130 pKa = 11.84APCRR134 pKa = 11.84KK135 pKa = 10.06CIDD138 pKa = 3.53RR139 pKa = 11.84EE140 pKa = 4.05II141 pKa = 5.35
Molecular weight: 16.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2377 |
98 |
597 |
339.6 |
38.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.721 ± 0.563 | 2.65 ± 0.928 |
7.068 ± 0.477 | 6.142 ± 0.535 |
4.544 ± 0.507 | 5.09 ± 0.725 |
2.188 ± 0.345 | 4.754 ± 0.794 |
5.469 ± 0.677 | 9.382 ± 0.546 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.43 ± 0.241 | 4.88 ± 0.392 |
6.31 ± 1.029 | 4.249 ± 0.591 |
5.848 ± 0.706 | 7.404 ± 0.449 |
5.89 ± 0.675 | 6.479 ± 0.787 |
1.262 ± 0.366 | 3.239 ± 0.333 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
