
Bovine faeces associated smacovirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus; Porprismacovirus bovas1
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
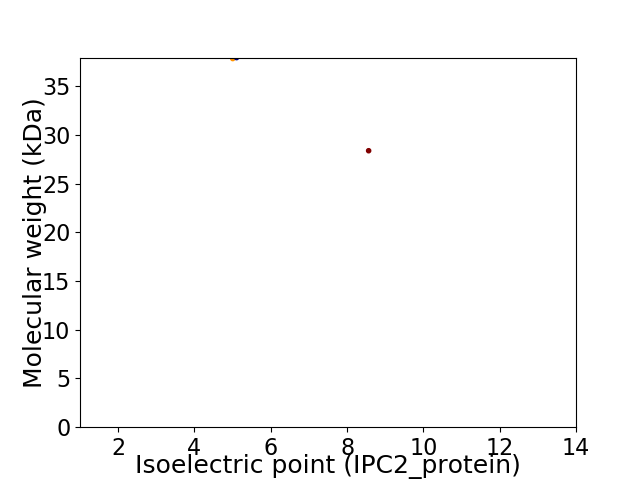
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
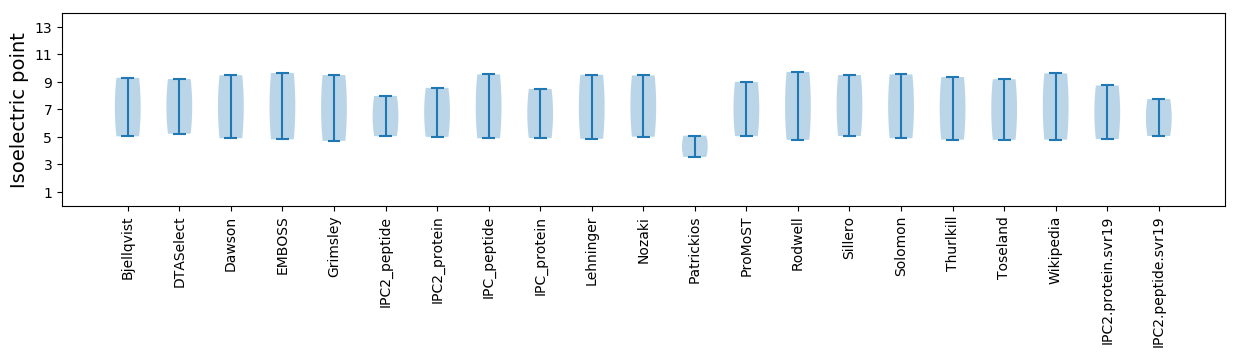
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A160HX09|A0A160HX09_9VIRU Replication associated protein OS=Bovine faeces associated smacovirus 2 OX=1843750 PE=4 SV=1
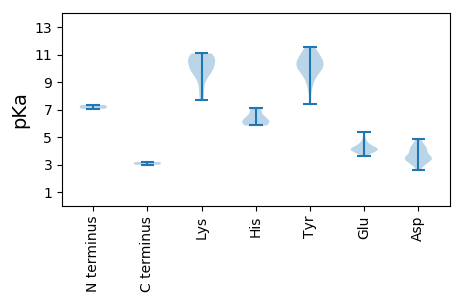
MM1 pKa = 7.06PTITVRR7 pKa = 11.84EE8 pKa = 4.18TYY10 pKa = 10.58DD11 pKa = 4.1LSTAPNRR18 pKa = 11.84LGLIGIHH25 pKa = 5.86TPGSAIIRR33 pKa = 11.84KK34 pKa = 9.17LYY36 pKa = 9.37PGLWLNYY43 pKa = 9.98NKK45 pKa = 10.14VKK47 pKa = 10.9VNSCNVALACASLLPADD64 pKa = 4.31PLQIGVEE71 pKa = 4.09AGDD74 pKa = 4.41IAPQDD79 pKa = 3.84LFNPILYY86 pKa = 8.83RR87 pKa = 11.84ACSTDD92 pKa = 2.89SFNTILNRR100 pKa = 11.84MYY102 pKa = 10.4GTDD105 pKa = 4.06SMFNLQDD112 pKa = 3.26GSIAFAGGTNPGDD125 pKa = 4.04PFPSASDD132 pKa = 3.49TVSEE136 pKa = 3.98DD137 pKa = 3.07VYY139 pKa = 11.53YY140 pKa = 11.26GLLSEE145 pKa = 5.11SGWKK149 pKa = 9.88KK150 pKa = 11.0AMPQSGLVMRR160 pKa = 11.84NLVPLVYY167 pKa = 10.51NVISNYY173 pKa = 10.59GNGAKK178 pKa = 10.02LDD180 pKa = 4.24GPSEE184 pKa = 4.26PNDD187 pKa = 3.45YY188 pKa = 10.02KK189 pKa = 11.06QVVAVPSGSGGSQSEE204 pKa = 4.12FRR206 pKa = 11.84LASIFRR212 pKa = 11.84GHH214 pKa = 6.51SFPMPALPTARR225 pKa = 11.84GLVQDD230 pKa = 3.56VVGYY234 pKa = 10.64QDD236 pKa = 3.59TSSSAQSTVDD246 pKa = 3.54NSWSFPYY253 pKa = 10.27FPPTLIPKK261 pKa = 8.91TYY263 pKa = 8.67VGCVIIPPSKK273 pKa = 9.93LHH275 pKa = 6.17RR276 pKa = 11.84LYY278 pKa = 11.12FRR280 pKa = 11.84MTISWSVSFFEE291 pKa = 5.38PISIVEE297 pKa = 4.18RR298 pKa = 11.84GRR300 pKa = 11.84LTEE303 pKa = 3.98ISQVGIATYY312 pKa = 10.23GSSYY316 pKa = 11.43NFGEE320 pKa = 4.79SKK322 pKa = 10.83DD323 pKa = 4.68LEE325 pKa = 4.67DD326 pKa = 4.13KK327 pKa = 9.88LTNIEE332 pKa = 4.34STVDD336 pKa = 3.41TLDD339 pKa = 3.27VDD341 pKa = 4.56AKK343 pKa = 10.65MIMQSS348 pKa = 2.98
MM1 pKa = 7.06PTITVRR7 pKa = 11.84EE8 pKa = 4.18TYY10 pKa = 10.58DD11 pKa = 4.1LSTAPNRR18 pKa = 11.84LGLIGIHH25 pKa = 5.86TPGSAIIRR33 pKa = 11.84KK34 pKa = 9.17LYY36 pKa = 9.37PGLWLNYY43 pKa = 9.98NKK45 pKa = 10.14VKK47 pKa = 10.9VNSCNVALACASLLPADD64 pKa = 4.31PLQIGVEE71 pKa = 4.09AGDD74 pKa = 4.41IAPQDD79 pKa = 3.84LFNPILYY86 pKa = 8.83RR87 pKa = 11.84ACSTDD92 pKa = 2.89SFNTILNRR100 pKa = 11.84MYY102 pKa = 10.4GTDD105 pKa = 4.06SMFNLQDD112 pKa = 3.26GSIAFAGGTNPGDD125 pKa = 4.04PFPSASDD132 pKa = 3.49TVSEE136 pKa = 3.98DD137 pKa = 3.07VYY139 pKa = 11.53YY140 pKa = 11.26GLLSEE145 pKa = 5.11SGWKK149 pKa = 9.88KK150 pKa = 11.0AMPQSGLVMRR160 pKa = 11.84NLVPLVYY167 pKa = 10.51NVISNYY173 pKa = 10.59GNGAKK178 pKa = 10.02LDD180 pKa = 4.24GPSEE184 pKa = 4.26PNDD187 pKa = 3.45YY188 pKa = 10.02KK189 pKa = 11.06QVVAVPSGSGGSQSEE204 pKa = 4.12FRR206 pKa = 11.84LASIFRR212 pKa = 11.84GHH214 pKa = 6.51SFPMPALPTARR225 pKa = 11.84GLVQDD230 pKa = 3.56VVGYY234 pKa = 10.64QDD236 pKa = 3.59TSSSAQSTVDD246 pKa = 3.54NSWSFPYY253 pKa = 10.27FPPTLIPKK261 pKa = 8.91TYY263 pKa = 8.67VGCVIIPPSKK273 pKa = 9.93LHH275 pKa = 6.17RR276 pKa = 11.84LYY278 pKa = 11.12FRR280 pKa = 11.84MTISWSVSFFEE291 pKa = 5.38PISIVEE297 pKa = 4.18RR298 pKa = 11.84GRR300 pKa = 11.84LTEE303 pKa = 3.98ISQVGIATYY312 pKa = 10.23GSSYY316 pKa = 11.43NFGEE320 pKa = 4.79SKK322 pKa = 10.83DD323 pKa = 4.68LEE325 pKa = 4.67DD326 pKa = 4.13KK327 pKa = 9.88LTNIEE332 pKa = 4.34STVDD336 pKa = 3.41TLDD339 pKa = 3.27VDD341 pKa = 4.56AKK343 pKa = 10.65MIMQSS348 pKa = 2.98
Molecular weight: 37.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A160HX09|A0A160HX09_9VIRU Replication associated protein OS=Bovine faeces associated smacovirus 2 OX=1843750 PE=4 SV=1
MM1 pKa = 7.33TGPRR5 pKa = 11.84TIHH8 pKa = 5.84KK9 pKa = 10.02RR10 pKa = 11.84IIRR13 pKa = 11.84EE14 pKa = 3.82IFKK17 pKa = 10.72KK18 pKa = 10.61CDD20 pKa = 3.08VKK22 pKa = 11.08KK23 pKa = 10.9YY24 pKa = 9.67IIAMEE29 pKa = 4.27TGNGGYY35 pKa = 7.41EE36 pKa = 3.68HH37 pKa = 6.35WQIRR41 pKa = 11.84CTASRR46 pKa = 11.84PDD48 pKa = 3.39FFEE51 pKa = 4.11YY52 pKa = 10.63VHH54 pKa = 7.13DD55 pKa = 4.15RR56 pKa = 11.84EE57 pKa = 4.5PRR59 pKa = 11.84FNIQKK64 pKa = 10.66AEE66 pKa = 4.14TEE68 pKa = 4.11SMEE71 pKa = 4.16YY72 pKa = 9.86EE73 pKa = 4.01RR74 pKa = 11.84KK75 pKa = 9.91EE76 pKa = 3.78GRR78 pKa = 11.84FWSSEE83 pKa = 3.62DD84 pKa = 3.25TTEE87 pKa = 4.32IRR89 pKa = 11.84QCRR92 pKa = 11.84FGEE95 pKa = 3.94LGGKK99 pKa = 8.34GRR101 pKa = 11.84EE102 pKa = 4.0WQRR105 pKa = 11.84QILKK109 pKa = 9.97VLKK112 pKa = 9.37NQDD115 pKa = 2.63VRR117 pKa = 11.84TIDD120 pKa = 3.5VVLDD124 pKa = 3.45PVGARR129 pKa = 11.84GKK131 pKa = 9.55SHH133 pKa = 6.11FTIALWEE140 pKa = 4.27RR141 pKa = 11.84GEE143 pKa = 3.99ALVVPRR149 pKa = 11.84YY150 pKa = 10.22SCTSEE155 pKa = 3.96KK156 pKa = 10.72LSAFVCSAYY165 pKa = 9.8KK166 pKa = 10.19GEE168 pKa = 4.57KK169 pKa = 9.55IIIIDD174 pKa = 3.53IPRR177 pKa = 11.84ANKK180 pKa = 7.67PTTALYY186 pKa = 10.95EE187 pKa = 4.44SMEE190 pKa = 4.02EE191 pKa = 4.1MKK193 pKa = 10.78DD194 pKa = 3.44GLVFDD199 pKa = 4.86PRR201 pKa = 11.84YY202 pKa = 9.88SGKK205 pKa = 8.15TRR207 pKa = 11.84NIRR210 pKa = 11.84GTKK213 pKa = 10.13VLVFTNNPLDD223 pKa = 4.06LKK225 pKa = 10.85KK226 pKa = 10.67LSHH229 pKa = 7.11DD230 pKa = 2.93RR231 pKa = 11.84WNLHH235 pKa = 6.02GITRR239 pKa = 11.84DD240 pKa = 3.51GTLSS244 pKa = 3.18
MM1 pKa = 7.33TGPRR5 pKa = 11.84TIHH8 pKa = 5.84KK9 pKa = 10.02RR10 pKa = 11.84IIRR13 pKa = 11.84EE14 pKa = 3.82IFKK17 pKa = 10.72KK18 pKa = 10.61CDD20 pKa = 3.08VKK22 pKa = 11.08KK23 pKa = 10.9YY24 pKa = 9.67IIAMEE29 pKa = 4.27TGNGGYY35 pKa = 7.41EE36 pKa = 3.68HH37 pKa = 6.35WQIRR41 pKa = 11.84CTASRR46 pKa = 11.84PDD48 pKa = 3.39FFEE51 pKa = 4.11YY52 pKa = 10.63VHH54 pKa = 7.13DD55 pKa = 4.15RR56 pKa = 11.84EE57 pKa = 4.5PRR59 pKa = 11.84FNIQKK64 pKa = 10.66AEE66 pKa = 4.14TEE68 pKa = 4.11SMEE71 pKa = 4.16YY72 pKa = 9.86EE73 pKa = 4.01RR74 pKa = 11.84KK75 pKa = 9.91EE76 pKa = 3.78GRR78 pKa = 11.84FWSSEE83 pKa = 3.62DD84 pKa = 3.25TTEE87 pKa = 4.32IRR89 pKa = 11.84QCRR92 pKa = 11.84FGEE95 pKa = 3.94LGGKK99 pKa = 8.34GRR101 pKa = 11.84EE102 pKa = 4.0WQRR105 pKa = 11.84QILKK109 pKa = 9.97VLKK112 pKa = 9.37NQDD115 pKa = 2.63VRR117 pKa = 11.84TIDD120 pKa = 3.5VVLDD124 pKa = 3.45PVGARR129 pKa = 11.84GKK131 pKa = 9.55SHH133 pKa = 6.11FTIALWEE140 pKa = 4.27RR141 pKa = 11.84GEE143 pKa = 3.99ALVVPRR149 pKa = 11.84YY150 pKa = 10.22SCTSEE155 pKa = 3.96KK156 pKa = 10.72LSAFVCSAYY165 pKa = 9.8KK166 pKa = 10.19GEE168 pKa = 4.57KK169 pKa = 9.55IIIIDD174 pKa = 3.53IPRR177 pKa = 11.84ANKK180 pKa = 7.67PTTALYY186 pKa = 10.95EE187 pKa = 4.44SMEE190 pKa = 4.02EE191 pKa = 4.1MKK193 pKa = 10.78DD194 pKa = 3.44GLVFDD199 pKa = 4.86PRR201 pKa = 11.84YY202 pKa = 9.88SGKK205 pKa = 8.15TRR207 pKa = 11.84NIRR210 pKa = 11.84GTKK213 pKa = 10.13VLVFTNNPLDD223 pKa = 4.06LKK225 pKa = 10.85KK226 pKa = 10.67LSHH229 pKa = 7.11DD230 pKa = 2.93RR231 pKa = 11.84WNLHH235 pKa = 6.02GITRR239 pKa = 11.84DD240 pKa = 3.51GTLSS244 pKa = 3.18
Molecular weight: 28.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
592 |
244 |
348 |
296.0 |
33.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.236 ± 0.655 | 1.52 ± 0.305 |
5.574 ± 0.142 | 5.574 ± 1.746 |
4.054 ± 0.026 | 7.77 ± 0.226 |
1.52 ± 0.541 | 6.926 ± 0.496 |
5.405 ± 1.608 | 7.601 ± 0.837 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.365 ± 0.182 | 4.392 ± 0.641 |
5.912 ± 1.281 | 2.872 ± 0.238 |
6.081 ± 1.927 | 8.784 ± 1.99 |
6.419 ± 0.316 | 6.419 ± 0.628 |
1.52 ± 0.305 | 4.054 ± 0.447 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
