
Puma concolor (Mountain lion)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Mammalia; Theria; Eutheria; Boreoeutheria; Laurasiatheria;
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
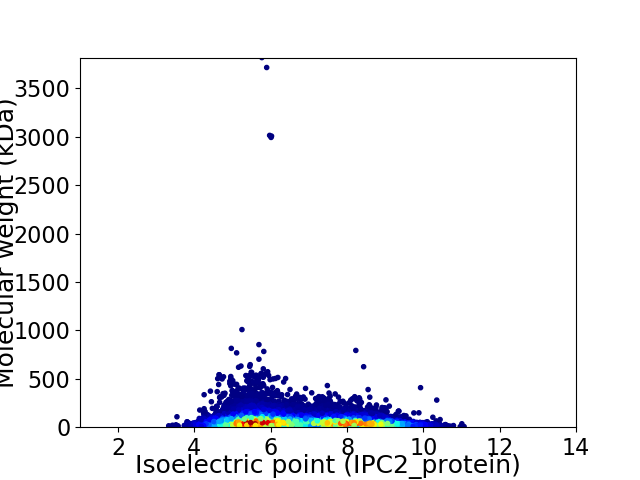
Virtual 2D-PAGE plot for 22836 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6P6HL58|A0A6P6HL58_PUMCO Isoform of A0A6P6HKZ6 contactin-associated protein-like 4 isoform X2 OS=Puma concolor OX=9696 GN=CNTNAP4 PE=3 SV=1
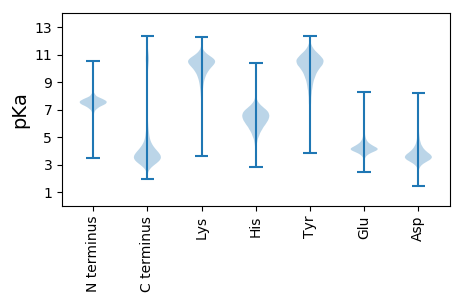
MM1 pKa = 7.22EE2 pKa = 5.09AVVLIPYY9 pKa = 9.99VLGLLLLPLLAVVLCVRR26 pKa = 11.84CRR28 pKa = 11.84EE29 pKa = 4.27LPGSYY34 pKa = 10.75DD35 pKa = 3.3NTASDD40 pKa = 3.94SLAPSSIVIKK50 pKa = 10.5RR51 pKa = 11.84PPTLATWTPATSYY64 pKa = 10.76PPVTSYY70 pKa = 11.28PPLSQPDD77 pKa = 4.23LLPIPRR83 pKa = 11.84SPQPPGGSHH92 pKa = 6.82RR93 pKa = 11.84MPSSRR98 pKa = 11.84QDD100 pKa = 2.92SDD102 pKa = 3.47GANSVASYY110 pKa = 10.11EE111 pKa = 4.13NEE113 pKa = 4.24GASGAPGALVAGRR126 pKa = 11.84LGPGLGPADD135 pKa = 3.58RR136 pKa = 11.84CVVTSEE142 pKa = 4.45PACEE146 pKa = 5.39DD147 pKa = 3.57DD148 pKa = 6.66DD149 pKa = 4.6EE150 pKa = 6.88DD151 pKa = 5.49EE152 pKa = 4.94EE153 pKa = 4.44EE154 pKa = 4.91DD155 pKa = 4.15YY156 pKa = 11.46PNEE159 pKa = 4.39GYY161 pKa = 11.04LEE163 pKa = 4.18VLPDD167 pKa = 3.64STPTTGTAVPPAPAPSNPGLRR188 pKa = 11.84DD189 pKa = 3.14SAFSMEE195 pKa = 4.23SGEE198 pKa = 4.99DD199 pKa = 3.63YY200 pKa = 11.66VNVPEE205 pKa = 4.61SEE207 pKa = 4.19EE208 pKa = 4.19SADD211 pKa = 3.6VSLDD215 pKa = 3.29GSRR218 pKa = 11.84EE219 pKa = 3.96YY220 pKa = 11.72VNVSQEE226 pKa = 4.24LPPVARR232 pKa = 11.84TEE234 pKa = 3.93PAILSSQNDD243 pKa = 3.92DD244 pKa = 4.02EE245 pKa = 5.46EE246 pKa = 5.21EE247 pKa = 4.2GAPDD251 pKa = 3.75YY252 pKa = 11.56EE253 pKa = 4.31NLQGLNN259 pKa = 3.55
MM1 pKa = 7.22EE2 pKa = 5.09AVVLIPYY9 pKa = 9.99VLGLLLLPLLAVVLCVRR26 pKa = 11.84CRR28 pKa = 11.84EE29 pKa = 4.27LPGSYY34 pKa = 10.75DD35 pKa = 3.3NTASDD40 pKa = 3.94SLAPSSIVIKK50 pKa = 10.5RR51 pKa = 11.84PPTLATWTPATSYY64 pKa = 10.76PPVTSYY70 pKa = 11.28PPLSQPDD77 pKa = 4.23LLPIPRR83 pKa = 11.84SPQPPGGSHH92 pKa = 6.82RR93 pKa = 11.84MPSSRR98 pKa = 11.84QDD100 pKa = 2.92SDD102 pKa = 3.47GANSVASYY110 pKa = 10.11EE111 pKa = 4.13NEE113 pKa = 4.24GASGAPGALVAGRR126 pKa = 11.84LGPGLGPADD135 pKa = 3.58RR136 pKa = 11.84CVVTSEE142 pKa = 4.45PACEE146 pKa = 5.39DD147 pKa = 3.57DD148 pKa = 6.66DD149 pKa = 4.6EE150 pKa = 6.88DD151 pKa = 5.49EE152 pKa = 4.94EE153 pKa = 4.44EE154 pKa = 4.91DD155 pKa = 4.15YY156 pKa = 11.46PNEE159 pKa = 4.39GYY161 pKa = 11.04LEE163 pKa = 4.18VLPDD167 pKa = 3.64STPTTGTAVPPAPAPSNPGLRR188 pKa = 11.84DD189 pKa = 3.14SAFSMEE195 pKa = 4.23SGEE198 pKa = 4.99DD199 pKa = 3.63YY200 pKa = 11.66VNVPEE205 pKa = 4.61SEE207 pKa = 4.19EE208 pKa = 4.19SADD211 pKa = 3.6VSLDD215 pKa = 3.29GSRR218 pKa = 11.84EE219 pKa = 3.96YY220 pKa = 11.72VNVSQEE226 pKa = 4.24LPPVARR232 pKa = 11.84TEE234 pKa = 3.93PAILSSQNDD243 pKa = 3.92DD244 pKa = 4.02EE245 pKa = 5.46EE246 pKa = 5.21EE247 pKa = 4.2GAPDD251 pKa = 3.75YY252 pKa = 11.56EE253 pKa = 4.31NLQGLNN259 pKa = 3.55
Molecular weight: 27.24 kDa
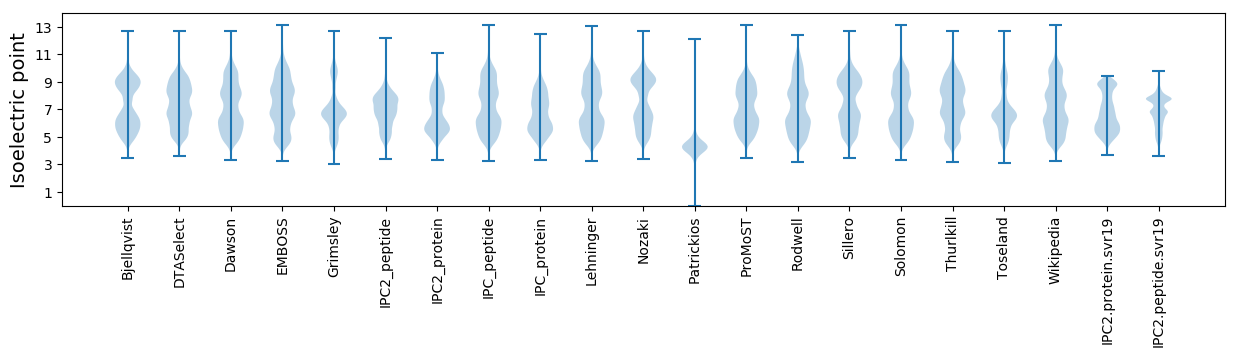
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P6HM51|A0A6P6HM51_PUMCO Thioredoxin-like protein OS=Puma concolor OX=9696 GN=TXNL4B PE=3 SV=1
MM1 pKa = 7.51GPGSGPNLRR10 pKa = 11.84AGVLLPSGNGPPNPRR25 pKa = 11.84PVGLGPGPSPNLRR38 pKa = 11.84SGFVGTNPAPRR49 pKa = 11.84SGMFPGPGLGPNPRR63 pKa = 11.84ANSLGPGLGPNPRR76 pKa = 11.84AGGLGPGPNLDD87 pKa = 3.45TRR89 pKa = 11.84AGGLLGTGSGLNLRR103 pKa = 11.84MAGPQGLDD111 pKa = 3.09LAPILRR117 pKa = 11.84AAGLLGANSAAFSQASGNMGTSPSSMARR145 pKa = 11.84VPGPMGPNSGPGSRR159 pKa = 11.84GIGLPGPNPSPMSRR173 pKa = 11.84APGPIGPNSAHH184 pKa = 7.19FSRR187 pKa = 11.84PAGPMGVNASPFPRR201 pKa = 11.84GTGSGGLNPAAFSQSSGTLASNPATFQRR229 pKa = 11.84SAGLQGSSPAIFPRR243 pKa = 11.84TSGPLGPNPANFPRR257 pKa = 11.84ASGLQGPSPAAFPRR271 pKa = 11.84STGPLGPGQVTFPRR285 pKa = 11.84SAPGPLGSSPAGPVGINPAPFARR308 pKa = 11.84PTGTLGLNPASFPRR322 pKa = 11.84MNGPVSKK329 pKa = 9.32TLVPFPRR336 pKa = 11.84VGNLPGTNPAAFPRR350 pKa = 11.84PGGPMAAMYY359 pKa = 9.22PNGMLPPP366 pKa = 4.73
MM1 pKa = 7.51GPGSGPNLRR10 pKa = 11.84AGVLLPSGNGPPNPRR25 pKa = 11.84PVGLGPGPSPNLRR38 pKa = 11.84SGFVGTNPAPRR49 pKa = 11.84SGMFPGPGLGPNPRR63 pKa = 11.84ANSLGPGLGPNPRR76 pKa = 11.84AGGLGPGPNLDD87 pKa = 3.45TRR89 pKa = 11.84AGGLLGTGSGLNLRR103 pKa = 11.84MAGPQGLDD111 pKa = 3.09LAPILRR117 pKa = 11.84AAGLLGANSAAFSQASGNMGTSPSSMARR145 pKa = 11.84VPGPMGPNSGPGSRR159 pKa = 11.84GIGLPGPNPSPMSRR173 pKa = 11.84APGPIGPNSAHH184 pKa = 7.19FSRR187 pKa = 11.84PAGPMGVNASPFPRR201 pKa = 11.84GTGSGGLNPAAFSQSSGTLASNPATFQRR229 pKa = 11.84SAGLQGSSPAIFPRR243 pKa = 11.84TSGPLGPNPANFPRR257 pKa = 11.84ASGLQGPSPAAFPRR271 pKa = 11.84STGPLGPGQVTFPRR285 pKa = 11.84SAPGPLGSSPAGPVGINPAPFARR308 pKa = 11.84PTGTLGLNPASFPRR322 pKa = 11.84MNGPVSKK329 pKa = 9.32TLVPFPRR336 pKa = 11.84VGNLPGTNPAAFPRR350 pKa = 11.84PGGPMAAMYY359 pKa = 9.22PNGMLPPP366 pKa = 4.73
Molecular weight: 35.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
13162948 |
31 |
34365 |
576.4 |
64.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.857 ± 0.016 | 2.194 ± 0.015 |
4.837 ± 0.011 | 7.114 ± 0.023 |
3.691 ± 0.013 | 6.517 ± 0.023 |
2.57 ± 0.01 | 4.481 ± 0.016 |
5.812 ± 0.025 | 9.887 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.174 ± 0.008 | 3.659 ± 0.012 |
6.174 ± 0.022 | 4.699 ± 0.016 |
5.664 ± 0.017 | 8.312 ± 0.025 |
5.35 ± 0.018 | 6.117 ± 0.018 |
1.194 ± 0.006 | 2.675 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
