
Pseudocercospora eumusae
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Dothideomycetidae; Mycosphaerellales; Mycosphaerellaceae; Pseudocercospora
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
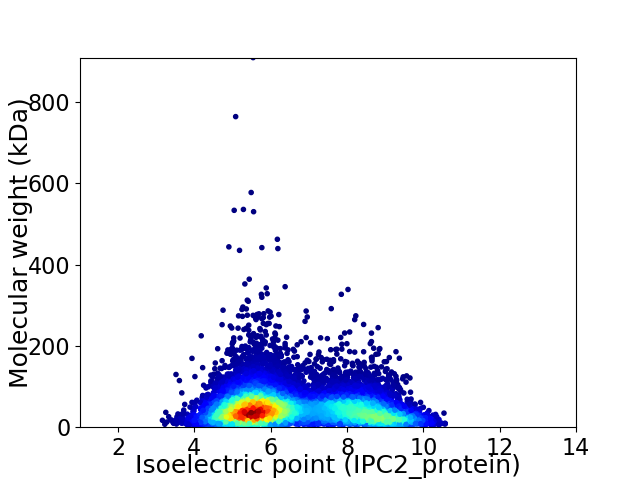
Virtual 2D-PAGE plot for 11956 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A139HHU4|A0A139HHU4_9PEZI Uncharacterized protein OS=Pseudocercospora eumusae OX=321146 GN=AC578_6514 PE=4 SV=1
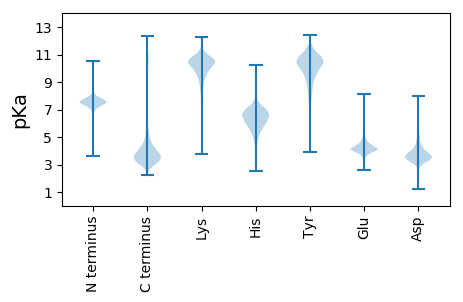
MM1 pKa = 8.1DD2 pKa = 3.6EE3 pKa = 4.72TYY5 pKa = 11.24SHH7 pKa = 7.46DD8 pKa = 3.98LVALAVKK15 pKa = 9.43MYY17 pKa = 9.75NSKK20 pKa = 10.62IIFAVLAAAIAVVRR34 pKa = 11.84SEE36 pKa = 4.44DD37 pKa = 4.11LEE39 pKa = 4.72PNDD42 pKa = 3.98VPPEE46 pKa = 4.12CRR48 pKa = 11.84SICQPAIDD56 pKa = 5.25LEE58 pKa = 4.35NSCDD62 pKa = 3.71RR63 pKa = 11.84QFDD66 pKa = 5.06DD67 pKa = 6.16DD68 pKa = 6.45DD69 pKa = 4.89NDD71 pKa = 4.03NNNSDD76 pKa = 3.28QQYY79 pKa = 10.35RR80 pKa = 11.84DD81 pKa = 4.26CVCKK85 pKa = 10.54AQNANQALSACQNCVVQYY103 pKa = 10.1PNYY106 pKa = 10.52FIDD109 pKa = 5.58ADD111 pKa = 5.3DD112 pKa = 5.46NDD114 pKa = 4.21TDD116 pKa = 5.21DD117 pKa = 5.14NDD119 pKa = 3.57IEE121 pKa = 4.24EE122 pKa = 4.35WLRR125 pKa = 11.84DD126 pKa = 3.9CGFSTATNQTGTAGVTPSPTRR147 pKa = 11.84GPGATGIFSGSLTTATYY164 pKa = 9.05TYY166 pKa = 10.85TEE168 pKa = 4.72IDD170 pKa = 4.71DD171 pKa = 6.02DD172 pKa = 6.51DD173 pKa = 7.46DD174 pKa = 6.33DD175 pKa = 5.01DD176 pKa = 4.71QPEE179 pKa = 4.48TEE181 pKa = 4.06TRR183 pKa = 11.84TTVFPVGAWTSGTFTGSYY201 pKa = 10.93GSLTTSTATYY211 pKa = 7.9TVPDD215 pKa = 5.37DD216 pKa = 6.44DD217 pKa = 7.33DD218 pKa = 7.26DD219 pKa = 7.56DD220 pKa = 6.46DD221 pKa = 6.92DD222 pKa = 5.56NDD224 pKa = 4.1SDD226 pKa = 4.39TITRR230 pKa = 11.84TLVFPAGATPSVPGQNTVSPTTGSVSFTTGTTTDD264 pKa = 3.18AQGNTEE270 pKa = 3.84VRR272 pKa = 11.84LRR274 pKa = 11.84LQDD277 pKa = 3.87LFLVPVWLSCWRR289 pKa = 11.84AWLYY293 pKa = 10.88RR294 pKa = 11.84GIVCTSKK301 pKa = 10.82RR302 pKa = 11.84LSCGLGLDD310 pKa = 3.36ICSGRR315 pKa = 11.84GRR317 pKa = 11.84LL318 pKa = 3.59
MM1 pKa = 8.1DD2 pKa = 3.6EE3 pKa = 4.72TYY5 pKa = 11.24SHH7 pKa = 7.46DD8 pKa = 3.98LVALAVKK15 pKa = 9.43MYY17 pKa = 9.75NSKK20 pKa = 10.62IIFAVLAAAIAVVRR34 pKa = 11.84SEE36 pKa = 4.44DD37 pKa = 4.11LEE39 pKa = 4.72PNDD42 pKa = 3.98VPPEE46 pKa = 4.12CRR48 pKa = 11.84SICQPAIDD56 pKa = 5.25LEE58 pKa = 4.35NSCDD62 pKa = 3.71RR63 pKa = 11.84QFDD66 pKa = 5.06DD67 pKa = 6.16DD68 pKa = 6.45DD69 pKa = 4.89NDD71 pKa = 4.03NNNSDD76 pKa = 3.28QQYY79 pKa = 10.35RR80 pKa = 11.84DD81 pKa = 4.26CVCKK85 pKa = 10.54AQNANQALSACQNCVVQYY103 pKa = 10.1PNYY106 pKa = 10.52FIDD109 pKa = 5.58ADD111 pKa = 5.3DD112 pKa = 5.46NDD114 pKa = 4.21TDD116 pKa = 5.21DD117 pKa = 5.14NDD119 pKa = 3.57IEE121 pKa = 4.24EE122 pKa = 4.35WLRR125 pKa = 11.84DD126 pKa = 3.9CGFSTATNQTGTAGVTPSPTRR147 pKa = 11.84GPGATGIFSGSLTTATYY164 pKa = 9.05TYY166 pKa = 10.85TEE168 pKa = 4.72IDD170 pKa = 4.71DD171 pKa = 6.02DD172 pKa = 6.51DD173 pKa = 7.46DD174 pKa = 6.33DD175 pKa = 5.01DD176 pKa = 4.71QPEE179 pKa = 4.48TEE181 pKa = 4.06TRR183 pKa = 11.84TTVFPVGAWTSGTFTGSYY201 pKa = 10.93GSLTTSTATYY211 pKa = 7.9TVPDD215 pKa = 5.37DD216 pKa = 6.44DD217 pKa = 7.33DD218 pKa = 7.26DD219 pKa = 7.56DD220 pKa = 6.46DD221 pKa = 6.92DD222 pKa = 5.56NDD224 pKa = 4.1SDD226 pKa = 4.39TITRR230 pKa = 11.84TLVFPAGATPSVPGQNTVSPTTGSVSFTTGTTTDD264 pKa = 3.18AQGNTEE270 pKa = 3.84VRR272 pKa = 11.84LRR274 pKa = 11.84LQDD277 pKa = 3.87LFLVPVWLSCWRR289 pKa = 11.84AWLYY293 pKa = 10.88RR294 pKa = 11.84GIVCTSKK301 pKa = 10.82RR302 pKa = 11.84LSCGLGLDD310 pKa = 3.36ICSGRR315 pKa = 11.84GRR317 pKa = 11.84LL318 pKa = 3.59
Molecular weight: 34.51 kDa
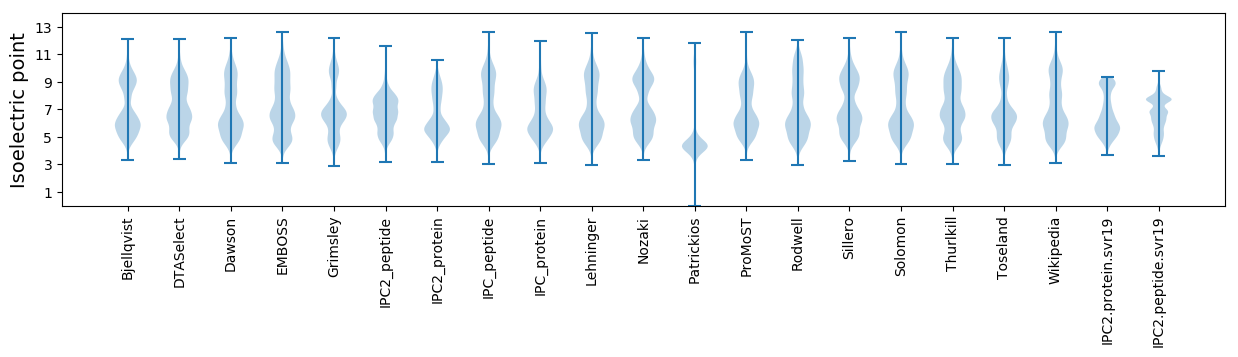
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A139GWA0|A0A139GWA0_9PEZI Isoform of A0A139GW97 60S ribosomal protein L36 OS=Pseudocercospora eumusae OX=321146 GN=AC578_1714 PE=3 SV=1
MM1 pKa = 7.25GQPSHH6 pKa = 6.97PASNALIYY14 pKa = 10.39CSYY17 pKa = 11.37AAFLVFGLFIAWRR30 pKa = 11.84LRR32 pKa = 11.84GQCAGEE38 pKa = 3.79WLSANRR44 pKa = 11.84TQKK47 pKa = 10.93AFPLALNFIASGEE60 pKa = 4.21FGLDD64 pKa = 2.89APRR67 pKa = 11.84IIVSSGRR74 pKa = 11.84RR75 pKa = 11.84QAGWKK80 pKa = 10.13CPEE83 pKa = 4.22TTSSNTKK90 pKa = 9.71VRR92 pKa = 11.84THH94 pKa = 7.02AFRR97 pKa = 11.84DD98 pKa = 3.43IALSAMILGSFARR111 pKa = 11.84RR112 pKa = 11.84VMCSTAGAKK121 pKa = 10.48GNILQVLCSRR131 pKa = 11.84YY132 pKa = 8.52LTDD135 pKa = 3.77CCTKK139 pKa = 10.41LRR141 pKa = 11.84RR142 pKa = 11.84TAMTRR147 pKa = 11.84SRR149 pKa = 3.84
MM1 pKa = 7.25GQPSHH6 pKa = 6.97PASNALIYY14 pKa = 10.39CSYY17 pKa = 11.37AAFLVFGLFIAWRR30 pKa = 11.84LRR32 pKa = 11.84GQCAGEE38 pKa = 3.79WLSANRR44 pKa = 11.84TQKK47 pKa = 10.93AFPLALNFIASGEE60 pKa = 4.21FGLDD64 pKa = 2.89APRR67 pKa = 11.84IIVSSGRR74 pKa = 11.84RR75 pKa = 11.84QAGWKK80 pKa = 10.13CPEE83 pKa = 4.22TTSSNTKK90 pKa = 9.71VRR92 pKa = 11.84THH94 pKa = 7.02AFRR97 pKa = 11.84DD98 pKa = 3.43IALSAMILGSFARR111 pKa = 11.84RR112 pKa = 11.84VMCSTAGAKK121 pKa = 10.48GNILQVLCSRR131 pKa = 11.84YY132 pKa = 8.52LTDD135 pKa = 3.77CCTKK139 pKa = 10.41LRR141 pKa = 11.84RR142 pKa = 11.84TAMTRR147 pKa = 11.84SRR149 pKa = 3.84
Molecular weight: 16.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5580759 |
34 |
8267 |
466.8 |
51.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.072 ± 0.02 | 1.265 ± 0.008 |
5.749 ± 0.015 | 6.196 ± 0.023 |
3.584 ± 0.013 | 6.722 ± 0.022 |
2.556 ± 0.01 | 4.725 ± 0.015 |
5.005 ± 0.018 | 8.584 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.233 ± 0.008 | 3.699 ± 0.013 |
6.0 ± 0.025 | 4.304 ± 0.016 |
6.188 ± 0.02 | 8.109 ± 0.026 |
5.971 ± 0.016 | 5.802 ± 0.017 |
1.47 ± 0.007 | 2.764 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
