
Acanthamoeba polyphaga mimivirus (APMV)
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Megaviricetes; Imitervirales; Mimiviridae; Mimivirus
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
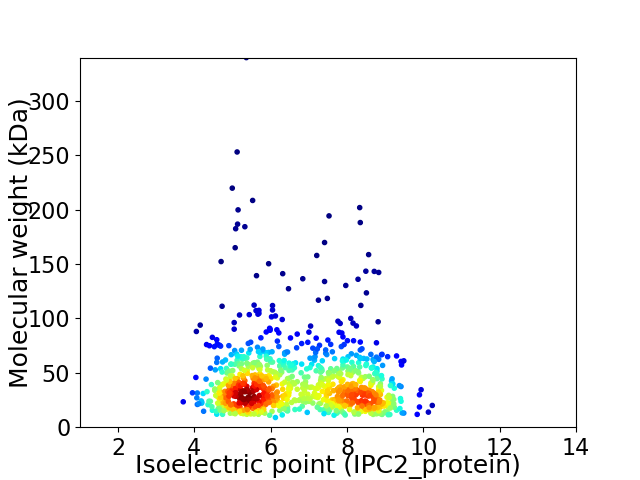
Virtual 2D-PAGE plot for 909 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
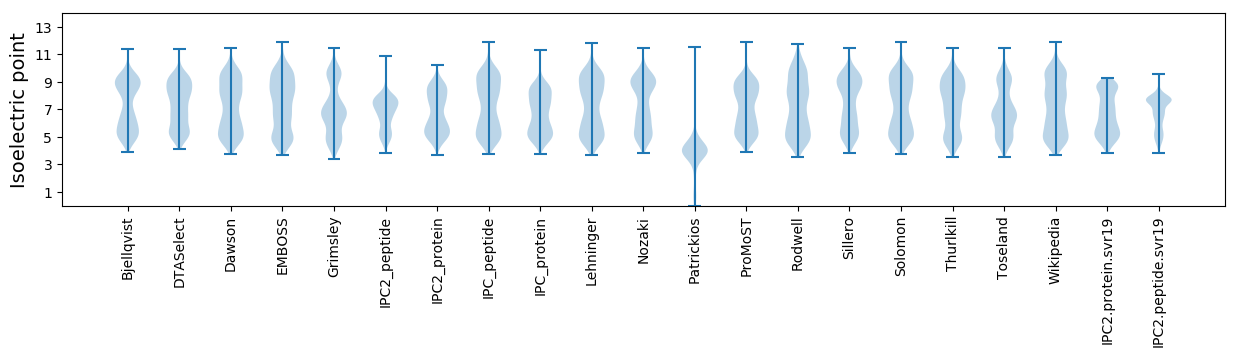
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q5UPV1|YL271_MIMIV Putative ankyrin repeat protein L271 OS=Acanthamoeba polyphaga mimivirus OX=212035 GN=MIMI_L271 PE=4 SV=1
MM1 pKa = 8.18DD2 pKa = 4.5FNKK5 pKa = 10.56LYY7 pKa = 11.17VLAQNNLFTDD17 pKa = 3.54ITIILKK23 pKa = 10.6DD24 pKa = 3.76EE25 pKa = 4.59SNEE28 pKa = 3.65ITLNLHH34 pKa = 6.0KK35 pKa = 10.82NIIYY39 pKa = 9.68SSCIFFEE46 pKa = 4.41KK47 pKa = 10.67LLTSFKK53 pKa = 10.39EE54 pKa = 4.14KK55 pKa = 10.26EE56 pKa = 3.94SSKK59 pKa = 10.74IILNVPNAIIVNDD72 pKa = 4.98IIWDD76 pKa = 4.44FYY78 pKa = 8.44GQKK81 pKa = 9.64IKK83 pKa = 10.65SHH85 pKa = 6.8NYY87 pKa = 8.99PEE89 pKa = 3.68WKK91 pKa = 10.51YY92 pKa = 10.62FIEE95 pKa = 4.47SYY97 pKa = 10.97KK98 pKa = 10.89CFDD101 pKa = 3.8YY102 pKa = 11.31FGMNTDD108 pKa = 3.05KK109 pKa = 11.22KK110 pKa = 10.54KK111 pKa = 10.41LYY113 pKa = 10.29KK114 pKa = 10.64LIVEE118 pKa = 4.31PDD120 pKa = 3.53GFDD123 pKa = 3.31EE124 pKa = 6.12LMDD127 pKa = 4.92FIDD130 pKa = 5.95LIGYY134 pKa = 9.07DD135 pKa = 3.65EE136 pKa = 4.4NAIEE140 pKa = 4.97VIFNNLPDD148 pKa = 5.5DD149 pKa = 4.32YY150 pKa = 11.28DD151 pKa = 3.66LSKK154 pKa = 10.82FSHH157 pKa = 6.21EE158 pKa = 4.99LLTEE162 pKa = 3.95MLSMYY167 pKa = 10.24KK168 pKa = 10.27KK169 pKa = 10.48NLVQIITRR177 pKa = 11.84NEE179 pKa = 3.86IIVLNDD185 pKa = 3.35FFDD188 pKa = 3.62GTIRR192 pKa = 11.84STEE195 pKa = 3.74HH196 pKa = 6.98CINSPEE202 pKa = 4.23VILFCASNASKK213 pKa = 10.48IVTFDD218 pKa = 3.13GHH220 pKa = 6.68NIRR223 pKa = 11.84ILEE226 pKa = 4.29SNNSILLSDD235 pKa = 4.85DD236 pKa = 3.54STVSDD241 pKa = 4.33AGTILHH247 pKa = 7.03ISHH250 pKa = 6.34SQNKK254 pKa = 8.09NVLVILYY261 pKa = 7.6EE262 pKa = 3.85YY263 pKa = 10.39RR264 pKa = 11.84IDD266 pKa = 3.55VWDD269 pKa = 3.6VLINKK274 pKa = 8.84LIASFRR280 pKa = 11.84VPFIKK285 pKa = 10.33KK286 pKa = 10.17LSCSYY291 pKa = 10.94DD292 pKa = 3.18GSQLVYY298 pKa = 10.87VDD300 pKa = 4.45NNNNMIVKK308 pKa = 10.18NILSEE313 pKa = 4.33EE314 pKa = 4.21IISEE318 pKa = 4.05INLQNLPEE326 pKa = 4.32ISNNFCIPEE335 pKa = 5.12LILEE339 pKa = 4.56TNTQDD344 pKa = 4.97LSEE347 pKa = 4.74IPNNLYY353 pKa = 9.03TLEE356 pKa = 4.67LKK358 pKa = 10.48SEE360 pKa = 4.45PDD362 pKa = 3.22QSIDD366 pKa = 3.86FIKK369 pKa = 11.13SNFQHH374 pKa = 6.64NLDD377 pKa = 4.93NLDD380 pKa = 4.67DD381 pKa = 5.41LNNLDD386 pKa = 5.45NSDD389 pKa = 5.28DD390 pKa = 4.14LDD392 pKa = 4.05NSNDD396 pKa = 3.74LNDD399 pKa = 5.08SNDD402 pKa = 4.08LDD404 pKa = 5.83DD405 pKa = 6.12SDD407 pKa = 6.19DD408 pKa = 4.53SNDD411 pKa = 3.84YY412 pKa = 11.72NNLNYY417 pKa = 10.52DD418 pKa = 4.1LNNLEE423 pKa = 5.25IICSSNNEE431 pKa = 3.46IFEE434 pKa = 4.42NVSTHH439 pKa = 5.17YY440 pKa = 10.96CIDD443 pKa = 3.7VNDD446 pKa = 4.81CNHH449 pKa = 6.78IDD451 pKa = 4.51NINSLNLQSPIQDD464 pKa = 3.86LSDD467 pKa = 3.45SLSPIDD473 pKa = 5.52YY474 pKa = 9.86IDD476 pKa = 3.83SSNLQSPVQDD486 pKa = 4.22LSDD489 pKa = 3.99SQSSTDD495 pKa = 3.66WIVSPNTQSPVYY507 pKa = 9.95YY508 pKa = 10.41LSKK511 pKa = 10.91LDD513 pKa = 3.69GSINGEE519 pKa = 4.16KK520 pKa = 10.6LSDD523 pKa = 3.55ALSIEE528 pKa = 4.19NDD530 pKa = 4.01SISDD534 pKa = 3.97NSDD537 pKa = 3.18NLNNSDD543 pKa = 5.2NSDD546 pKa = 4.39DD547 pKa = 5.53LDD549 pKa = 4.73NPDD552 pKa = 4.9NSDD555 pKa = 3.56NLDD558 pKa = 3.54NSDD561 pKa = 3.89YY562 pKa = 11.16LLEE565 pKa = 4.12YY566 pKa = 10.53LVNTDD571 pKa = 2.65KK572 pKa = 11.2TNYY575 pKa = 8.72MFNLTYY581 pKa = 10.45SIHH584 pKa = 5.86TDD586 pKa = 2.83KK587 pKa = 11.13TNKK590 pKa = 9.68RR591 pKa = 11.84FNLEE595 pKa = 3.65NPYY598 pKa = 10.74EE599 pKa = 4.03QFNFINKK606 pKa = 9.17FCLLYY611 pKa = 10.57INNKK615 pKa = 9.02VIYY618 pKa = 9.62FIDD621 pKa = 4.86IIDD624 pKa = 3.56KK625 pKa = 10.76KK626 pKa = 10.31IIHH629 pKa = 6.74KK630 pKa = 7.16EE631 pKa = 3.78TCRR634 pKa = 11.84RR635 pKa = 11.84NIVNMRR641 pKa = 11.84CSPTGDD647 pKa = 3.5SIAIIDD653 pKa = 3.87SEE655 pKa = 5.07YY656 pKa = 10.44IVYY659 pKa = 9.93IYY661 pKa = 11.16KK662 pKa = 10.68LVTDD666 pKa = 4.21SNGFLCYY673 pKa = 9.98PIVDD677 pKa = 3.62YY678 pKa = 11.22CILVRR683 pKa = 11.84DD684 pKa = 4.63LFLPTKK690 pKa = 10.16IEE692 pKa = 3.94YY693 pKa = 10.24SSNGRR698 pKa = 11.84YY699 pKa = 9.17LVFDD703 pKa = 5.04DD704 pKa = 5.99DD705 pKa = 4.38GLSINLYY712 pKa = 10.41DD713 pKa = 4.26IEE715 pKa = 4.12QLEE718 pKa = 4.32MTNEE722 pKa = 3.86FEE724 pKa = 5.04LFCDD728 pKa = 3.39SKK730 pKa = 11.13IIVDD734 pKa = 3.31ITFSEE739 pKa = 4.91NYY741 pKa = 9.41CDD743 pKa = 4.25EE744 pKa = 5.22LINRR748 pKa = 11.84LNNALKK754 pKa = 10.31KK755 pKa = 10.15IEE757 pKa = 4.17QKK759 pKa = 10.98YY760 pKa = 9.64PNN762 pKa = 3.88
MM1 pKa = 8.18DD2 pKa = 4.5FNKK5 pKa = 10.56LYY7 pKa = 11.17VLAQNNLFTDD17 pKa = 3.54ITIILKK23 pKa = 10.6DD24 pKa = 3.76EE25 pKa = 4.59SNEE28 pKa = 3.65ITLNLHH34 pKa = 6.0KK35 pKa = 10.82NIIYY39 pKa = 9.68SSCIFFEE46 pKa = 4.41KK47 pKa = 10.67LLTSFKK53 pKa = 10.39EE54 pKa = 4.14KK55 pKa = 10.26EE56 pKa = 3.94SSKK59 pKa = 10.74IILNVPNAIIVNDD72 pKa = 4.98IIWDD76 pKa = 4.44FYY78 pKa = 8.44GQKK81 pKa = 9.64IKK83 pKa = 10.65SHH85 pKa = 6.8NYY87 pKa = 8.99PEE89 pKa = 3.68WKK91 pKa = 10.51YY92 pKa = 10.62FIEE95 pKa = 4.47SYY97 pKa = 10.97KK98 pKa = 10.89CFDD101 pKa = 3.8YY102 pKa = 11.31FGMNTDD108 pKa = 3.05KK109 pKa = 11.22KK110 pKa = 10.54KK111 pKa = 10.41LYY113 pKa = 10.29KK114 pKa = 10.64LIVEE118 pKa = 4.31PDD120 pKa = 3.53GFDD123 pKa = 3.31EE124 pKa = 6.12LMDD127 pKa = 4.92FIDD130 pKa = 5.95LIGYY134 pKa = 9.07DD135 pKa = 3.65EE136 pKa = 4.4NAIEE140 pKa = 4.97VIFNNLPDD148 pKa = 5.5DD149 pKa = 4.32YY150 pKa = 11.28DD151 pKa = 3.66LSKK154 pKa = 10.82FSHH157 pKa = 6.21EE158 pKa = 4.99LLTEE162 pKa = 3.95MLSMYY167 pKa = 10.24KK168 pKa = 10.27KK169 pKa = 10.48NLVQIITRR177 pKa = 11.84NEE179 pKa = 3.86IIVLNDD185 pKa = 3.35FFDD188 pKa = 3.62GTIRR192 pKa = 11.84STEE195 pKa = 3.74HH196 pKa = 6.98CINSPEE202 pKa = 4.23VILFCASNASKK213 pKa = 10.48IVTFDD218 pKa = 3.13GHH220 pKa = 6.68NIRR223 pKa = 11.84ILEE226 pKa = 4.29SNNSILLSDD235 pKa = 4.85DD236 pKa = 3.54STVSDD241 pKa = 4.33AGTILHH247 pKa = 7.03ISHH250 pKa = 6.34SQNKK254 pKa = 8.09NVLVILYY261 pKa = 7.6EE262 pKa = 3.85YY263 pKa = 10.39RR264 pKa = 11.84IDD266 pKa = 3.55VWDD269 pKa = 3.6VLINKK274 pKa = 8.84LIASFRR280 pKa = 11.84VPFIKK285 pKa = 10.33KK286 pKa = 10.17LSCSYY291 pKa = 10.94DD292 pKa = 3.18GSQLVYY298 pKa = 10.87VDD300 pKa = 4.45NNNNMIVKK308 pKa = 10.18NILSEE313 pKa = 4.33EE314 pKa = 4.21IISEE318 pKa = 4.05INLQNLPEE326 pKa = 4.32ISNNFCIPEE335 pKa = 5.12LILEE339 pKa = 4.56TNTQDD344 pKa = 4.97LSEE347 pKa = 4.74IPNNLYY353 pKa = 9.03TLEE356 pKa = 4.67LKK358 pKa = 10.48SEE360 pKa = 4.45PDD362 pKa = 3.22QSIDD366 pKa = 3.86FIKK369 pKa = 11.13SNFQHH374 pKa = 6.64NLDD377 pKa = 4.93NLDD380 pKa = 4.67DD381 pKa = 5.41LNNLDD386 pKa = 5.45NSDD389 pKa = 5.28DD390 pKa = 4.14LDD392 pKa = 4.05NSNDD396 pKa = 3.74LNDD399 pKa = 5.08SNDD402 pKa = 4.08LDD404 pKa = 5.83DD405 pKa = 6.12SDD407 pKa = 6.19DD408 pKa = 4.53SNDD411 pKa = 3.84YY412 pKa = 11.72NNLNYY417 pKa = 10.52DD418 pKa = 4.1LNNLEE423 pKa = 5.25IICSSNNEE431 pKa = 3.46IFEE434 pKa = 4.42NVSTHH439 pKa = 5.17YY440 pKa = 10.96CIDD443 pKa = 3.7VNDD446 pKa = 4.81CNHH449 pKa = 6.78IDD451 pKa = 4.51NINSLNLQSPIQDD464 pKa = 3.86LSDD467 pKa = 3.45SLSPIDD473 pKa = 5.52YY474 pKa = 9.86IDD476 pKa = 3.83SSNLQSPVQDD486 pKa = 4.22LSDD489 pKa = 3.99SQSSTDD495 pKa = 3.66WIVSPNTQSPVYY507 pKa = 9.95YY508 pKa = 10.41LSKK511 pKa = 10.91LDD513 pKa = 3.69GSINGEE519 pKa = 4.16KK520 pKa = 10.6LSDD523 pKa = 3.55ALSIEE528 pKa = 4.19NDD530 pKa = 4.01SISDD534 pKa = 3.97NSDD537 pKa = 3.18NLNNSDD543 pKa = 5.2NSDD546 pKa = 4.39DD547 pKa = 5.53LDD549 pKa = 4.73NPDD552 pKa = 4.9NSDD555 pKa = 3.56NLDD558 pKa = 3.54NSDD561 pKa = 3.89YY562 pKa = 11.16LLEE565 pKa = 4.12YY566 pKa = 10.53LVNTDD571 pKa = 2.65KK572 pKa = 11.2TNYY575 pKa = 8.72MFNLTYY581 pKa = 10.45SIHH584 pKa = 5.86TDD586 pKa = 2.83KK587 pKa = 11.13TNKK590 pKa = 9.68RR591 pKa = 11.84FNLEE595 pKa = 3.65NPYY598 pKa = 10.74EE599 pKa = 4.03QFNFINKK606 pKa = 9.17FCLLYY611 pKa = 10.57INNKK615 pKa = 9.02VIYY618 pKa = 9.62FIDD621 pKa = 4.86IIDD624 pKa = 3.56KK625 pKa = 10.76KK626 pKa = 10.31IIHH629 pKa = 6.74KK630 pKa = 7.16EE631 pKa = 3.78TCRR634 pKa = 11.84RR635 pKa = 11.84NIVNMRR641 pKa = 11.84CSPTGDD647 pKa = 3.5SIAIIDD653 pKa = 3.87SEE655 pKa = 5.07YY656 pKa = 10.44IVYY659 pKa = 9.93IYY661 pKa = 11.16KK662 pKa = 10.68LVTDD666 pKa = 4.21SNGFLCYY673 pKa = 9.98PIVDD677 pKa = 3.62YY678 pKa = 11.22CILVRR683 pKa = 11.84DD684 pKa = 4.63LFLPTKK690 pKa = 10.16IEE692 pKa = 3.94YY693 pKa = 10.24SSNGRR698 pKa = 11.84YY699 pKa = 9.17LVFDD703 pKa = 5.04DD704 pKa = 5.99DD705 pKa = 4.38GLSINLYY712 pKa = 10.41DD713 pKa = 4.26IEE715 pKa = 4.12QLEE718 pKa = 4.32MTNEE722 pKa = 3.86FEE724 pKa = 5.04LFCDD728 pKa = 3.39SKK730 pKa = 11.13IIVDD734 pKa = 3.31ITFSEE739 pKa = 4.91NYY741 pKa = 9.41CDD743 pKa = 4.25EE744 pKa = 5.22LINRR748 pKa = 11.84LNNALKK754 pKa = 10.31KK755 pKa = 10.15IEE757 pKa = 4.17QKK759 pKa = 10.98YY760 pKa = 9.64PNN762 pKa = 3.88
Molecular weight: 88.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q5UQ00|FPG_MIMIV Probable formamidopyrimidine-DNA glycosylase OS=Acanthamoeba polyphaga mimivirus OX=212035 GN=MIMI_L315 PE=1 SV=3
MM1 pKa = 7.52TSLNTNVEE9 pKa = 4.05HH10 pKa = 7.12RR11 pKa = 11.84RR12 pKa = 11.84TRR14 pKa = 11.84TTISQIIHH22 pKa = 7.38DD23 pKa = 4.46MTPEE27 pKa = 3.6NKK29 pKa = 9.9FYY31 pKa = 10.09ITVADD36 pKa = 4.08GGSKK40 pKa = 10.27RR41 pKa = 11.84FRR43 pKa = 11.84NKK45 pKa = 10.21PNDD48 pKa = 3.44QSRR51 pKa = 11.84HH52 pKa = 4.74SGQYY56 pKa = 8.27QPRR59 pKa = 11.84NLSGKK64 pKa = 7.56TNISTQSQFTPRR76 pKa = 11.84LYY78 pKa = 10.72GKK80 pKa = 9.76QYY82 pKa = 11.23NYY84 pKa = 10.35NQPNRR89 pKa = 11.84LQTRR93 pKa = 11.84SVRR96 pKa = 11.84PNYY99 pKa = 10.2YY100 pKa = 9.75SGYY103 pKa = 9.33RR104 pKa = 11.84YY105 pKa = 10.58NNRR108 pKa = 11.84TGNQEE113 pKa = 4.69FYY115 pKa = 11.02PMQPFNNQSFNNQSRR130 pKa = 11.84THH132 pKa = 5.28QSKK135 pKa = 8.71TYY137 pKa = 9.02QHH139 pKa = 5.97NQQKK143 pKa = 10.51RR144 pKa = 11.84SFNGPRR150 pKa = 11.84NNGPQNNVPRR160 pKa = 11.84FFQRR164 pKa = 11.84EE165 pKa = 4.0EE166 pKa = 4.23TNN168 pKa = 3.15
MM1 pKa = 7.52TSLNTNVEE9 pKa = 4.05HH10 pKa = 7.12RR11 pKa = 11.84RR12 pKa = 11.84TRR14 pKa = 11.84TTISQIIHH22 pKa = 7.38DD23 pKa = 4.46MTPEE27 pKa = 3.6NKK29 pKa = 9.9FYY31 pKa = 10.09ITVADD36 pKa = 4.08GGSKK40 pKa = 10.27RR41 pKa = 11.84FRR43 pKa = 11.84NKK45 pKa = 10.21PNDD48 pKa = 3.44QSRR51 pKa = 11.84HH52 pKa = 4.74SGQYY56 pKa = 8.27QPRR59 pKa = 11.84NLSGKK64 pKa = 7.56TNISTQSQFTPRR76 pKa = 11.84LYY78 pKa = 10.72GKK80 pKa = 9.76QYY82 pKa = 11.23NYY84 pKa = 10.35NQPNRR89 pKa = 11.84LQTRR93 pKa = 11.84SVRR96 pKa = 11.84PNYY99 pKa = 10.2YY100 pKa = 9.75SGYY103 pKa = 9.33RR104 pKa = 11.84YY105 pKa = 10.58NNRR108 pKa = 11.84TGNQEE113 pKa = 4.69FYY115 pKa = 11.02PMQPFNNQSFNNQSRR130 pKa = 11.84THH132 pKa = 5.28QSKK135 pKa = 8.71TYY137 pKa = 9.02QHH139 pKa = 5.97NQQKK143 pKa = 10.51RR144 pKa = 11.84SFNGPRR150 pKa = 11.84NNGPQNNVPRR160 pKa = 11.84FFQRR164 pKa = 11.84EE165 pKa = 4.0EE166 pKa = 4.23TNN168 pKa = 3.15
Molecular weight: 20.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
341649 |
81 |
2959 |
375.9 |
43.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.072 ± 0.062 | 1.801 ± 0.048 |
6.795 ± 0.071 | 5.493 ± 0.072 |
4.617 ± 0.059 | 4.642 ± 0.216 |
2.054 ± 0.036 | 9.86 ± 0.101 |
8.981 ± 0.128 | 8.16 ± 0.087 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.124 ± 0.033 | 8.883 ± 0.101 |
3.225 ± 0.059 | 3.137 ± 0.056 |
3.292 ± 0.056 | 7.382 ± 0.072 |
5.253 ± 0.056 | 5.054 ± 0.062 |
0.738 ± 0.022 | 5.435 ± 0.06 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
