
Arthrobacter sp. MN05-02
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Micrococcaceae; Arthrobacter; unclassified Arthrobacter
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
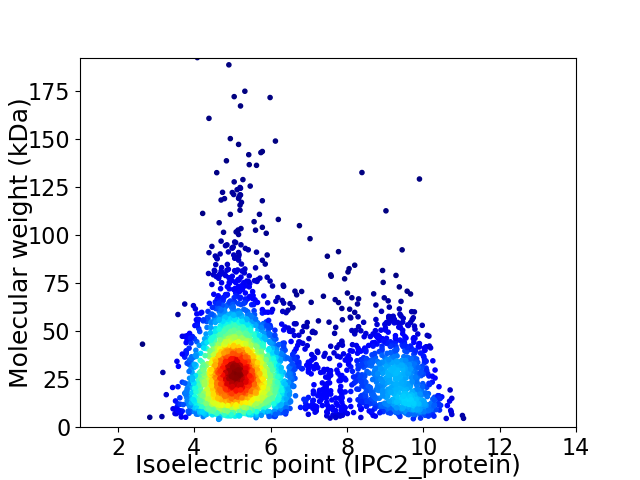
Virtual 2D-PAGE plot for 3539 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q9WF81|A0A3Q9WF81_9MICC Arginine biosynthesis bifunctional protein ArgJ OS=Arthrobacter sp. MN05-02 OX=1571833 GN=argJ PE=3 SV=1
MM1 pKa = 7.49KK2 pKa = 10.44SKK4 pKa = 10.49ISPAVLRR11 pKa = 11.84GSALALAAALALTACGGGGNDD32 pKa = 4.51AGGAAPAEE40 pKa = 4.35VSADD44 pKa = 3.72DD45 pKa = 3.78VQAALDD51 pKa = 3.66KK52 pKa = 11.66GGDD55 pKa = 3.54LTVWAWEE62 pKa = 3.88PTLEE66 pKa = 4.61KK67 pKa = 10.7VVEE70 pKa = 4.3DD71 pKa = 5.72FEE73 pKa = 6.28AEE75 pKa = 4.14HH76 pKa = 6.8PNVNIDD82 pKa = 3.93LVNAGTGNDD91 pKa = 3.59QYY93 pKa = 11.47TALQNAIAAGSGVPDD108 pKa = 3.67VAQIEE113 pKa = 5.14YY114 pKa = 10.6YY115 pKa = 10.86ALPQFALAEE124 pKa = 4.36SVQDD128 pKa = 3.55LSGYY132 pKa = 9.91GAGDD136 pKa = 3.53LQDD139 pKa = 3.85TFSPGPWSSVQANEE153 pKa = 4.67GIYY156 pKa = 10.01GLPMDD161 pKa = 5.41SGPMALFYY169 pKa = 10.15NTEE172 pKa = 3.86VFEE175 pKa = 4.68KK176 pKa = 10.88YY177 pKa = 10.2GVEE180 pKa = 4.98VPTTWEE186 pKa = 4.24EE187 pKa = 3.94YY188 pKa = 10.55VEE190 pKa = 4.2AARR193 pKa = 11.84TLHH196 pKa = 6.37EE197 pKa = 5.37ADD199 pKa = 3.56PDD201 pKa = 3.99VYY203 pKa = 10.59IANDD207 pKa = 3.3TGDD210 pKa = 3.73AGFATSLIWQAGGQPYY226 pKa = 8.28TVDD229 pKa = 3.36GTDD232 pKa = 3.13VKK234 pKa = 11.43VDD236 pKa = 4.17FSDD239 pKa = 3.52EE240 pKa = 4.19GSTKK244 pKa = 10.05FAEE247 pKa = 4.5TWQQLLDD254 pKa = 4.14EE255 pKa = 5.53DD256 pKa = 4.49LLAPISSWSDD266 pKa = 2.5EE267 pKa = 4.18WYY269 pKa = 10.44QGLGTGTIATLATGAWMPGNFTSGVPAAAGKK300 pKa = 8.54WRR302 pKa = 11.84AAPLPQWEE310 pKa = 4.5EE311 pKa = 4.02GGTASAEE318 pKa = 4.15NGGSSLTIPAASEE331 pKa = 3.84QGALAYY337 pKa = 10.48AFLDD341 pKa = 3.83YY342 pKa = 11.48ANVGDD347 pKa = 4.54GVQTRR352 pKa = 11.84IDD354 pKa = 3.57GGAFPATSAALEE366 pKa = 4.0SDD368 pKa = 3.46EE369 pKa = 4.29FLNSEE374 pKa = 4.22FEE376 pKa = 4.32YY377 pKa = 11.03FGGQKK382 pKa = 10.45ANEE385 pKa = 4.08IFAEE389 pKa = 4.39SAQNVPEE396 pKa = 4.39GWSYY400 pKa = 11.82LPFQVYY406 pKa = 10.66ANSIFNDD413 pKa = 3.97TVGQAYY419 pKa = 10.45VSDD422 pKa = 4.07TPLKK426 pKa = 10.82DD427 pKa = 4.23GLKK430 pKa = 9.18SWQDD434 pKa = 3.03ASIKK438 pKa = 10.8YY439 pKa = 8.32GTEE442 pKa = 3.36QGFTVSEE449 pKa = 4.18
MM1 pKa = 7.49KK2 pKa = 10.44SKK4 pKa = 10.49ISPAVLRR11 pKa = 11.84GSALALAAALALTACGGGGNDD32 pKa = 4.51AGGAAPAEE40 pKa = 4.35VSADD44 pKa = 3.72DD45 pKa = 3.78VQAALDD51 pKa = 3.66KK52 pKa = 11.66GGDD55 pKa = 3.54LTVWAWEE62 pKa = 3.88PTLEE66 pKa = 4.61KK67 pKa = 10.7VVEE70 pKa = 4.3DD71 pKa = 5.72FEE73 pKa = 6.28AEE75 pKa = 4.14HH76 pKa = 6.8PNVNIDD82 pKa = 3.93LVNAGTGNDD91 pKa = 3.59QYY93 pKa = 11.47TALQNAIAAGSGVPDD108 pKa = 3.67VAQIEE113 pKa = 5.14YY114 pKa = 10.6YY115 pKa = 10.86ALPQFALAEE124 pKa = 4.36SVQDD128 pKa = 3.55LSGYY132 pKa = 9.91GAGDD136 pKa = 3.53LQDD139 pKa = 3.85TFSPGPWSSVQANEE153 pKa = 4.67GIYY156 pKa = 10.01GLPMDD161 pKa = 5.41SGPMALFYY169 pKa = 10.15NTEE172 pKa = 3.86VFEE175 pKa = 4.68KK176 pKa = 10.88YY177 pKa = 10.2GVEE180 pKa = 4.98VPTTWEE186 pKa = 4.24EE187 pKa = 3.94YY188 pKa = 10.55VEE190 pKa = 4.2AARR193 pKa = 11.84TLHH196 pKa = 6.37EE197 pKa = 5.37ADD199 pKa = 3.56PDD201 pKa = 3.99VYY203 pKa = 10.59IANDD207 pKa = 3.3TGDD210 pKa = 3.73AGFATSLIWQAGGQPYY226 pKa = 8.28TVDD229 pKa = 3.36GTDD232 pKa = 3.13VKK234 pKa = 11.43VDD236 pKa = 4.17FSDD239 pKa = 3.52EE240 pKa = 4.19GSTKK244 pKa = 10.05FAEE247 pKa = 4.5TWQQLLDD254 pKa = 4.14EE255 pKa = 5.53DD256 pKa = 4.49LLAPISSWSDD266 pKa = 2.5EE267 pKa = 4.18WYY269 pKa = 10.44QGLGTGTIATLATGAWMPGNFTSGVPAAAGKK300 pKa = 8.54WRR302 pKa = 11.84AAPLPQWEE310 pKa = 4.5EE311 pKa = 4.02GGTASAEE318 pKa = 4.15NGGSSLTIPAASEE331 pKa = 3.84QGALAYY337 pKa = 10.48AFLDD341 pKa = 3.83YY342 pKa = 11.48ANVGDD347 pKa = 4.54GVQTRR352 pKa = 11.84IDD354 pKa = 3.57GGAFPATSAALEE366 pKa = 4.0SDD368 pKa = 3.46EE369 pKa = 4.29FLNSEE374 pKa = 4.22FEE376 pKa = 4.32YY377 pKa = 11.03FGGQKK382 pKa = 10.45ANEE385 pKa = 4.08IFAEE389 pKa = 4.39SAQNVPEE396 pKa = 4.39GWSYY400 pKa = 11.82LPFQVYY406 pKa = 10.66ANSIFNDD413 pKa = 3.97TVGQAYY419 pKa = 10.45VSDD422 pKa = 4.07TPLKK426 pKa = 10.82DD427 pKa = 4.23GLKK430 pKa = 9.18SWQDD434 pKa = 3.03ASIKK438 pKa = 10.8YY439 pKa = 8.32GTEE442 pKa = 3.36QGFTVSEE449 pKa = 4.18
Molecular weight: 47.24 kDa
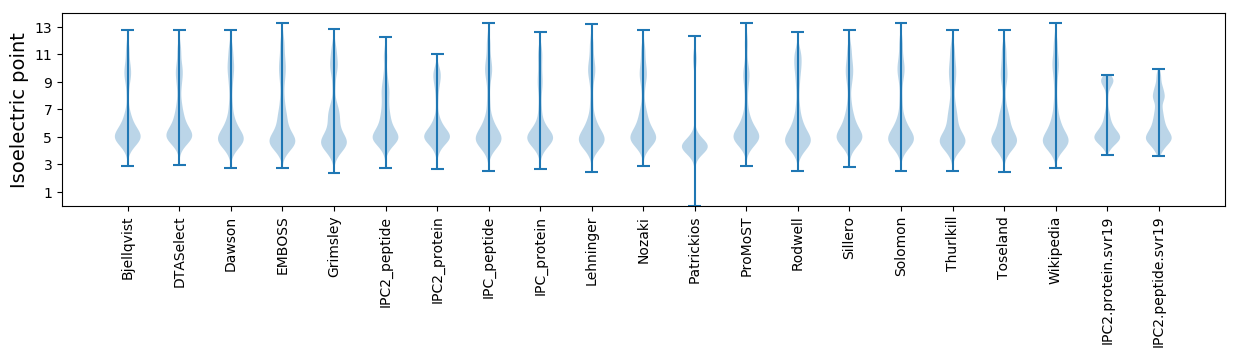
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3T0Y8M2|A0A3T0Y8M2_9MICC Coenzyme A pyrophosphatase OS=Arthrobacter sp. MN05-02 OX=1571833 GN=MN0502_27990 PE=4 SV=1
MM1 pKa = 7.61KK2 pKa = 10.31VRR4 pKa = 11.84NSLRR8 pKa = 11.84ALKK11 pKa = 9.65KK12 pKa = 10.17IPGAQIVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GKK24 pKa = 8.33TLVINKK30 pKa = 9.19KK31 pKa = 8.38NPRR34 pKa = 11.84MKK36 pKa = 10.47ARR38 pKa = 11.84QGG40 pKa = 3.24
MM1 pKa = 7.61KK2 pKa = 10.31VRR4 pKa = 11.84NSLRR8 pKa = 11.84ALKK11 pKa = 9.65KK12 pKa = 10.17IPGAQIVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GKK24 pKa = 8.33TLVINKK30 pKa = 9.19KK31 pKa = 8.38NPRR34 pKa = 11.84MKK36 pKa = 10.47ARR38 pKa = 11.84QGG40 pKa = 3.24
Molecular weight: 4.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1059819 |
39 |
1826 |
299.5 |
32.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.957 ± 0.052 | 0.607 ± 0.01 |
6.101 ± 0.036 | 5.621 ± 0.039 |
3.076 ± 0.023 | 9.265 ± 0.038 |
2.093 ± 0.021 | 4.121 ± 0.033 |
2.028 ± 0.026 | 10.261 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.826 ± 0.016 | 2.08 ± 0.023 |
5.645 ± 0.034 | 3.017 ± 0.022 |
7.358 ± 0.047 | 5.889 ± 0.027 |
6.091 ± 0.037 | 8.574 ± 0.037 |
1.371 ± 0.016 | 2.019 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
