
Branchiostoma belcheri (Amphioxus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Cephalochordata; Leptocardii; Amphioxiformes; Branchiostomidae; Branchiostoma
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
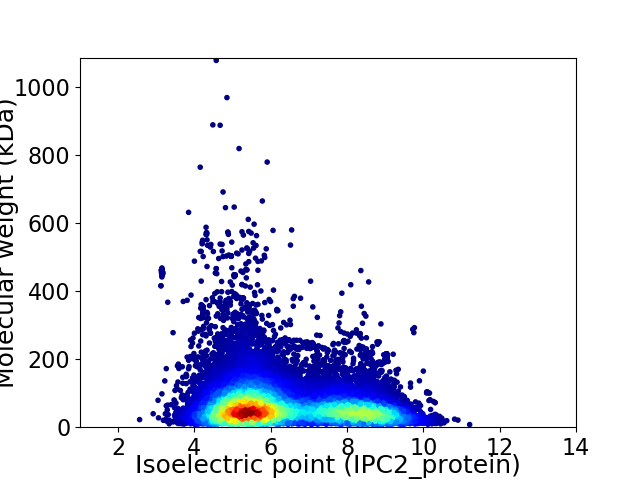
Virtual 2D-PAGE plot for 31614 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6P4ZVL0|A0A6P4ZVL0_BRABE probable G-protein coupled receptor 139 OS=Branchiostoma belcheri OX=7741 GN=LOC109480299 PE=4 SV=1
MM1 pKa = 7.3TPILLFFVAVATAAPMRR18 pKa = 11.84RR19 pKa = 11.84QTRR22 pKa = 11.84ILDD25 pKa = 3.69ANGDD29 pKa = 3.97GLISKK34 pKa = 10.1PEE36 pKa = 3.94VTSVMTLHH44 pKa = 7.2DD45 pKa = 4.67ALVALDD51 pKa = 3.44IDD53 pKa = 4.47GDD55 pKa = 3.98HH56 pKa = 7.04FLYY59 pKa = 10.62LPQIVEE65 pKa = 4.15LFGDD69 pKa = 4.31GSIFYY74 pKa = 10.42LLNSNGDD81 pKa = 3.46DD82 pKa = 3.18HH83 pKa = 8.81LSFGEE88 pKa = 4.28VQHH91 pKa = 6.62GLTLSEE97 pKa = 4.94FFDD100 pKa = 5.15LFDD103 pKa = 4.68KK104 pKa = 11.56NGDD107 pKa = 3.68GVLDD111 pKa = 3.81TSEE114 pKa = 4.57SYY116 pKa = 10.57QMNYY120 pKa = 9.53IYY122 pKa = 10.04NTIQNPDD129 pKa = 3.33AAITNALDD137 pKa = 3.58ANGDD141 pKa = 3.72GKK143 pKa = 10.94LSKK146 pKa = 11.09LEE148 pKa = 3.86VLGAMTLEE156 pKa = 4.12EE157 pKa = 4.79SMIAMDD163 pKa = 4.59ADD165 pKa = 3.57GDD167 pKa = 4.26GFFTVQEE174 pKa = 4.06LMPIFGDD181 pKa = 3.62VTQATSDD188 pKa = 3.63QLDD191 pKa = 3.67TNMDD195 pKa = 3.61GLLSFDD201 pKa = 3.48EE202 pKa = 4.73VTFGTSLDD210 pKa = 4.24RR211 pKa = 11.84IFDD214 pKa = 4.35FYY216 pKa = 11.77DD217 pKa = 3.92LDD219 pKa = 5.3GDD221 pKa = 4.43GYY223 pKa = 10.05LTGSEE228 pKa = 4.27ADD230 pKa = 3.84GIYY233 pKa = 10.47YY234 pKa = 10.39VYY236 pKa = 10.01TAIVNNPILVHH247 pKa = 6.55GGLDD251 pKa = 3.43ADD253 pKa = 4.31GDD255 pKa = 4.45GKK257 pKa = 11.21LSLPEE262 pKa = 3.92VEE264 pKa = 4.5LAMNIHH270 pKa = 7.06DD271 pKa = 5.33ALLALDD277 pKa = 3.75QDD279 pKa = 4.54GDD281 pKa = 3.54NHH283 pKa = 7.48LNEE286 pKa = 4.33QEE288 pKa = 4.12FVHH291 pKa = 6.19YY292 pKa = 10.97VGDD295 pKa = 3.41QLLFHH300 pKa = 6.52TLDD303 pKa = 3.56HH304 pKa = 6.89NGDD307 pKa = 3.34QTLSFGEE314 pKa = 4.11MHH316 pKa = 6.89NLGLDD321 pKa = 3.62TLFNQYY327 pKa = 10.61DD328 pKa = 3.72KK329 pKa = 11.72NGDD332 pKa = 3.49GFLEE336 pKa = 4.33GLEE339 pKa = 4.43AEE341 pKa = 4.75TMLHH345 pKa = 6.8VYY347 pKa = 10.79DD348 pKa = 5.47LVQAMNAAA356 pKa = 3.88
MM1 pKa = 7.3TPILLFFVAVATAAPMRR18 pKa = 11.84RR19 pKa = 11.84QTRR22 pKa = 11.84ILDD25 pKa = 3.69ANGDD29 pKa = 3.97GLISKK34 pKa = 10.1PEE36 pKa = 3.94VTSVMTLHH44 pKa = 7.2DD45 pKa = 4.67ALVALDD51 pKa = 3.44IDD53 pKa = 4.47GDD55 pKa = 3.98HH56 pKa = 7.04FLYY59 pKa = 10.62LPQIVEE65 pKa = 4.15LFGDD69 pKa = 4.31GSIFYY74 pKa = 10.42LLNSNGDD81 pKa = 3.46DD82 pKa = 3.18HH83 pKa = 8.81LSFGEE88 pKa = 4.28VQHH91 pKa = 6.62GLTLSEE97 pKa = 4.94FFDD100 pKa = 5.15LFDD103 pKa = 4.68KK104 pKa = 11.56NGDD107 pKa = 3.68GVLDD111 pKa = 3.81TSEE114 pKa = 4.57SYY116 pKa = 10.57QMNYY120 pKa = 9.53IYY122 pKa = 10.04NTIQNPDD129 pKa = 3.33AAITNALDD137 pKa = 3.58ANGDD141 pKa = 3.72GKK143 pKa = 10.94LSKK146 pKa = 11.09LEE148 pKa = 3.86VLGAMTLEE156 pKa = 4.12EE157 pKa = 4.79SMIAMDD163 pKa = 4.59ADD165 pKa = 3.57GDD167 pKa = 4.26GFFTVQEE174 pKa = 4.06LMPIFGDD181 pKa = 3.62VTQATSDD188 pKa = 3.63QLDD191 pKa = 3.67TNMDD195 pKa = 3.61GLLSFDD201 pKa = 3.48EE202 pKa = 4.73VTFGTSLDD210 pKa = 4.24RR211 pKa = 11.84IFDD214 pKa = 4.35FYY216 pKa = 11.77DD217 pKa = 3.92LDD219 pKa = 5.3GDD221 pKa = 4.43GYY223 pKa = 10.05LTGSEE228 pKa = 4.27ADD230 pKa = 3.84GIYY233 pKa = 10.47YY234 pKa = 10.39VYY236 pKa = 10.01TAIVNNPILVHH247 pKa = 6.55GGLDD251 pKa = 3.43ADD253 pKa = 4.31GDD255 pKa = 4.45GKK257 pKa = 11.21LSLPEE262 pKa = 3.92VEE264 pKa = 4.5LAMNIHH270 pKa = 7.06DD271 pKa = 5.33ALLALDD277 pKa = 3.75QDD279 pKa = 4.54GDD281 pKa = 3.54NHH283 pKa = 7.48LNEE286 pKa = 4.33QEE288 pKa = 4.12FVHH291 pKa = 6.19YY292 pKa = 10.97VGDD295 pKa = 3.41QLLFHH300 pKa = 6.52TLDD303 pKa = 3.56HH304 pKa = 6.89NGDD307 pKa = 3.34QTLSFGEE314 pKa = 4.11MHH316 pKa = 6.89NLGLDD321 pKa = 3.62TLFNQYY327 pKa = 10.61DD328 pKa = 3.72KK329 pKa = 11.72NGDD332 pKa = 3.49GFLEE336 pKa = 4.33GLEE339 pKa = 4.43AEE341 pKa = 4.75TMLHH345 pKa = 6.8VYY347 pKa = 10.79DD348 pKa = 5.47LVQAMNAAA356 pKa = 3.88
Molecular weight: 39.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P4YBX3|A0A6P4YBX3_BRABE uncharacterized protein LOC109463859 OS=Branchiostoma belcheri OX=7741 GN=LOC109463859 PE=4 SV=1
MM1 pKa = 7.85PKK3 pKa = 9.97RR4 pKa = 11.84ARR6 pKa = 11.84SASGSRR12 pKa = 11.84KK13 pKa = 8.78PAAKK17 pKa = 9.68KK18 pKa = 10.23AKK20 pKa = 9.4PAAKK24 pKa = 9.78RR25 pKa = 11.84KK26 pKa = 9.97APAAAAAAAPPAKK39 pKa = 9.99KK40 pKa = 10.1ARR42 pKa = 11.84PAKK45 pKa = 10.15AKK47 pKa = 9.89KK48 pKa = 10.24APRR51 pKa = 11.84KK52 pKa = 9.34KK53 pKa = 10.0AAKK56 pKa = 9.52PRR58 pKa = 11.84AKK60 pKa = 9.77PAKK63 pKa = 9.95KK64 pKa = 10.14SFKK67 pKa = 10.28KK68 pKa = 10.32KK69 pKa = 10.07AGGRR73 pKa = 11.84KK74 pKa = 8.99KK75 pKa = 10.69
MM1 pKa = 7.85PKK3 pKa = 9.97RR4 pKa = 11.84ARR6 pKa = 11.84SASGSRR12 pKa = 11.84KK13 pKa = 8.78PAAKK17 pKa = 9.68KK18 pKa = 10.23AKK20 pKa = 9.4PAAKK24 pKa = 9.78RR25 pKa = 11.84KK26 pKa = 9.97APAAAAAAAPPAKK39 pKa = 9.99KK40 pKa = 10.1ARR42 pKa = 11.84PAKK45 pKa = 10.15AKK47 pKa = 9.89KK48 pKa = 10.24APRR51 pKa = 11.84KK52 pKa = 9.34KK53 pKa = 10.0AAKK56 pKa = 9.52PRR58 pKa = 11.84AKK60 pKa = 9.77PAKK63 pKa = 9.95KK64 pKa = 10.14SFKK67 pKa = 10.28KK68 pKa = 10.32KK69 pKa = 10.07AGGRR73 pKa = 11.84KK74 pKa = 8.99KK75 pKa = 10.69
Molecular weight: 7.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
20550264 |
54 |
24062 |
650.0 |
72.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.859 ± 0.013 | 2.307 ± 0.022 |
5.962 ± 0.017 | 6.821 ± 0.021 |
3.316 ± 0.01 | 6.805 ± 0.023 |
2.411 ± 0.008 | 4.133 ± 0.01 |
5.46 ± 0.02 | 8.317 ± 0.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.381 ± 0.007 | 3.996 ± 0.012 |
5.733 ± 0.018 | 4.741 ± 0.018 |
5.631 ± 0.013 | 7.759 ± 0.015 |
6.753 ± 0.03 | 6.582 ± 0.012 |
1.175 ± 0.004 | 2.85 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
