
Jeotgalicoccus saudimassiliensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Staphylococcaceae; Jeotgalicoccus
Average proteome isoelectric point is 5.85
Get precalculated fractions of proteins
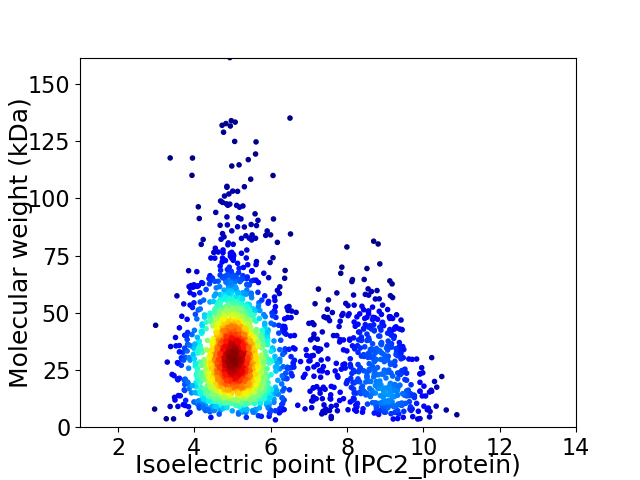
Virtual 2D-PAGE plot for 2181 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A078MED2|A0A078MED2_9STAP D-alanyl-D-alanine carboxypeptidase OS=Jeotgalicoccus saudimassiliensis OX=1461582 GN=vanY PE=4 SV=1
MM1 pKa = 6.65STKK4 pKa = 10.53NFLIMVGGILAVLFFLLALAIRR26 pKa = 11.84AEE28 pKa = 4.17MGGDD32 pKa = 3.5DD33 pKa = 4.07TGTAGKK39 pKa = 8.04TAGDD43 pKa = 3.64TEE45 pKa = 4.34EE46 pKa = 4.22QQTEE50 pKa = 4.42TEE52 pKa = 4.29SQEE55 pKa = 3.98TALNLNDD62 pKa = 3.58SPLYY66 pKa = 10.29AVHH69 pKa = 7.33EE70 pKa = 4.29DD71 pKa = 3.56TAPVFQYY78 pKa = 10.97QSGVDD83 pKa = 3.15IPYY86 pKa = 10.02PEE88 pKa = 4.69EE89 pKa = 4.14GVKK92 pKa = 10.55GIYY95 pKa = 7.99VTAYY99 pKa = 8.8SAGGEE104 pKa = 4.08RR105 pKa = 11.84MPEE108 pKa = 4.6LIDD111 pKa = 3.95LVNNTGLNSMVIDD124 pKa = 3.89VKK126 pKa = 11.23EE127 pKa = 4.59DD128 pKa = 2.87IGDD131 pKa = 3.15IMMPLDD137 pKa = 3.68VDD139 pKa = 3.43NDD141 pKa = 3.2IVRR144 pKa = 11.84NHH146 pKa = 6.35MYY148 pKa = 10.58DD149 pKa = 3.54YY150 pKa = 11.19VDD152 pKa = 4.32PKK154 pKa = 11.61ALMTTMEE161 pKa = 4.36EE162 pKa = 4.04NEE164 pKa = 4.39IYY166 pKa = 10.22PIARR170 pKa = 11.84IVVFKK175 pKa = 10.57DD176 pKa = 2.7SRR178 pKa = 11.84LAMEE182 pKa = 5.26RR183 pKa = 11.84PDD185 pKa = 4.91LSYY188 pKa = 11.31LNPDD192 pKa = 2.89GSVWQNGSGEE202 pKa = 4.41SFVNPFLKK210 pKa = 10.41EE211 pKa = 3.71VWDD214 pKa = 4.05YY215 pKa = 11.61NVDD218 pKa = 3.66VAIEE222 pKa = 3.81AAKK225 pKa = 10.72LGFKK229 pKa = 9.94EE230 pKa = 3.86IQFDD234 pKa = 3.86YY235 pKa = 11.38VRR237 pKa = 11.84FPEE240 pKa = 4.58SFDD243 pKa = 3.63TLSSGLTYY251 pKa = 11.02DD252 pKa = 3.4FGEE255 pKa = 4.5YY256 pKa = 10.89ANTEE260 pKa = 3.9ADD262 pKa = 3.37EE263 pKa = 4.23VQQRR267 pKa = 11.84VNAVTDD273 pKa = 4.36FVAYY277 pKa = 10.1ASEE280 pKa = 3.97QLKK283 pKa = 10.61PYY285 pKa = 10.5DD286 pKa = 3.32VDD288 pKa = 3.61VSVDD292 pKa = 3.43VFGYY296 pKa = 10.81AATQRR301 pKa = 11.84EE302 pKa = 4.44APGIGQNFSQIADD315 pKa = 3.56NVDD318 pKa = 3.9IISSMIYY325 pKa = 8.5PSHH328 pKa = 6.27WGAYY332 pKa = 9.46SFDD335 pKa = 3.15IAAPDD340 pKa = 3.76TEE342 pKa = 4.82PYY344 pKa = 10.95AVVDD348 pKa = 3.09QYY350 pKa = 11.61MKK352 pKa = 11.02VEE354 pKa = 3.98NEE356 pKa = 3.89VLGVLEE362 pKa = 4.99EE363 pKa = 4.52PPEE366 pKa = 4.12SRR368 pKa = 11.84PWIQDD373 pKa = 2.74FTAGDD378 pKa = 4.57LGPGNYY384 pKa = 8.56IEE386 pKa = 4.69YY387 pKa = 10.06NAPEE391 pKa = 4.25VEE393 pKa = 4.2AQIQALKK400 pKa = 11.1DD401 pKa = 3.58NGIDD405 pKa = 5.75EE406 pKa = 4.46YY407 pKa = 11.77LLWNAQNEE415 pKa = 4.39YY416 pKa = 11.03SEE418 pKa = 4.27GTEE421 pKa = 4.05YY422 pKa = 10.86QQ423 pKa = 3.2
MM1 pKa = 6.65STKK4 pKa = 10.53NFLIMVGGILAVLFFLLALAIRR26 pKa = 11.84AEE28 pKa = 4.17MGGDD32 pKa = 3.5DD33 pKa = 4.07TGTAGKK39 pKa = 8.04TAGDD43 pKa = 3.64TEE45 pKa = 4.34EE46 pKa = 4.22QQTEE50 pKa = 4.42TEE52 pKa = 4.29SQEE55 pKa = 3.98TALNLNDD62 pKa = 3.58SPLYY66 pKa = 10.29AVHH69 pKa = 7.33EE70 pKa = 4.29DD71 pKa = 3.56TAPVFQYY78 pKa = 10.97QSGVDD83 pKa = 3.15IPYY86 pKa = 10.02PEE88 pKa = 4.69EE89 pKa = 4.14GVKK92 pKa = 10.55GIYY95 pKa = 7.99VTAYY99 pKa = 8.8SAGGEE104 pKa = 4.08RR105 pKa = 11.84MPEE108 pKa = 4.6LIDD111 pKa = 3.95LVNNTGLNSMVIDD124 pKa = 3.89VKK126 pKa = 11.23EE127 pKa = 4.59DD128 pKa = 2.87IGDD131 pKa = 3.15IMMPLDD137 pKa = 3.68VDD139 pKa = 3.43NDD141 pKa = 3.2IVRR144 pKa = 11.84NHH146 pKa = 6.35MYY148 pKa = 10.58DD149 pKa = 3.54YY150 pKa = 11.19VDD152 pKa = 4.32PKK154 pKa = 11.61ALMTTMEE161 pKa = 4.36EE162 pKa = 4.04NEE164 pKa = 4.39IYY166 pKa = 10.22PIARR170 pKa = 11.84IVVFKK175 pKa = 10.57DD176 pKa = 2.7SRR178 pKa = 11.84LAMEE182 pKa = 5.26RR183 pKa = 11.84PDD185 pKa = 4.91LSYY188 pKa = 11.31LNPDD192 pKa = 2.89GSVWQNGSGEE202 pKa = 4.41SFVNPFLKK210 pKa = 10.41EE211 pKa = 3.71VWDD214 pKa = 4.05YY215 pKa = 11.61NVDD218 pKa = 3.66VAIEE222 pKa = 3.81AAKK225 pKa = 10.72LGFKK229 pKa = 9.94EE230 pKa = 3.86IQFDD234 pKa = 3.86YY235 pKa = 11.38VRR237 pKa = 11.84FPEE240 pKa = 4.58SFDD243 pKa = 3.63TLSSGLTYY251 pKa = 11.02DD252 pKa = 3.4FGEE255 pKa = 4.5YY256 pKa = 10.89ANTEE260 pKa = 3.9ADD262 pKa = 3.37EE263 pKa = 4.23VQQRR267 pKa = 11.84VNAVTDD273 pKa = 4.36FVAYY277 pKa = 10.1ASEE280 pKa = 3.97QLKK283 pKa = 10.61PYY285 pKa = 10.5DD286 pKa = 3.32VDD288 pKa = 3.61VSVDD292 pKa = 3.43VFGYY296 pKa = 10.81AATQRR301 pKa = 11.84EE302 pKa = 4.44APGIGQNFSQIADD315 pKa = 3.56NVDD318 pKa = 3.9IISSMIYY325 pKa = 8.5PSHH328 pKa = 6.27WGAYY332 pKa = 9.46SFDD335 pKa = 3.15IAAPDD340 pKa = 3.76TEE342 pKa = 4.82PYY344 pKa = 10.95AVVDD348 pKa = 3.09QYY350 pKa = 11.61MKK352 pKa = 11.02VEE354 pKa = 3.98NEE356 pKa = 3.89VLGVLEE362 pKa = 4.99EE363 pKa = 4.52PPEE366 pKa = 4.12SRR368 pKa = 11.84PWIQDD373 pKa = 2.74FTAGDD378 pKa = 4.57LGPGNYY384 pKa = 8.56IEE386 pKa = 4.69YY387 pKa = 10.06NAPEE391 pKa = 4.25VEE393 pKa = 4.2AQIQALKK400 pKa = 11.1DD401 pKa = 3.58NGIDD405 pKa = 5.75EE406 pKa = 4.46YY407 pKa = 11.77LLWNAQNEE415 pKa = 4.39YY416 pKa = 11.03SEE418 pKa = 4.27GTEE421 pKa = 4.05YY422 pKa = 10.86QQ423 pKa = 3.2
Molecular weight: 47.13 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A078MCC8|A0A078MCC8_9STAP YciI-like protein OS=Jeotgalicoccus saudimassiliensis OX=1461582 GN=BN1048_01969 PE=3 SV=1
MM1 pKa = 7.49VKK3 pKa = 9.1RR4 pKa = 11.84TYY6 pKa = 10.42QPNKK10 pKa = 8.33RR11 pKa = 11.84KK12 pKa = 9.38RR13 pKa = 11.84AKK15 pKa = 9.1VHH17 pKa = 5.5GFRR20 pKa = 11.84EE21 pKa = 4.12RR22 pKa = 11.84MSTKK26 pKa = 10.23NGRR29 pKa = 11.84KK30 pKa = 8.49VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.05GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
MM1 pKa = 7.49VKK3 pKa = 9.1RR4 pKa = 11.84TYY6 pKa = 10.42QPNKK10 pKa = 8.33RR11 pKa = 11.84KK12 pKa = 9.38RR13 pKa = 11.84AKK15 pKa = 9.1VHH17 pKa = 5.5GFRR20 pKa = 11.84EE21 pKa = 4.12RR22 pKa = 11.84MSTKK26 pKa = 10.23NGRR29 pKa = 11.84KK30 pKa = 8.49VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.05GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
Molecular weight: 5.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
655108 |
30 |
1427 |
300.4 |
33.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.081 ± 0.061 | 0.512 ± 0.013 |
6.035 ± 0.048 | 7.802 ± 0.07 |
4.56 ± 0.038 | 6.869 ± 0.054 |
2.076 ± 0.025 | 8.044 ± 0.058 |
6.312 ± 0.054 | 9.3 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.984 ± 0.024 | 4.868 ± 0.04 |
3.334 ± 0.028 | 2.956 ± 0.031 |
3.933 ± 0.045 | 5.972 ± 0.039 |
5.79 ± 0.028 | 7.074 ± 0.044 |
0.739 ± 0.014 | 3.76 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
